- Transcript BART1_0-u18690.013
- Transcript IDBART1_0-u18690.013
- Gene IDBART1_0-u18690
Exon Structure of BART1_0-u18690.013
| Chromosome | Exon | Start | Stop | Direction |
|---|---|---|---|---|
| chr3H | 1 | 67423171 | 67421743 | - |
| chr3H | 2 | 67425255 | 67425189 | - |
| chr3H | 3 | 67425680 | 67425530 | - |
| chr3H | 4 | 67425991 | 67425751 | - |
| chr3H | 5 | 67426259 | 67426201 | - |
| chr3H | 6 | 67426950 | 67426694 | - |
| chr3H | 7 | 67427189 | 67427047 | - |
| chr3H | 8 | 67427332 | 67427278 | - |
| chr3H | 9 | 67427506 | 67427429 | - |
| chr3H | 10 | 67427705 | 67427601 | - |
| chr3H | 11 | 67428995 | 67427855 | - |
grey: non-coding, green: Barley Morex IBSC 2017 Transcript CDS, red: Barley RTD exons
Salmon TPM Values: sixteensamplesmorex
These TPM values were generated by using the RNA-seq data from a 16-tissue experiment in Morex (published here) were calculated using Salmon (version Salmon-0.8.2) using BaRTv1.0-QUASI as the reference transcript dataset. Click here for more information about the RNA-seq experiment and materials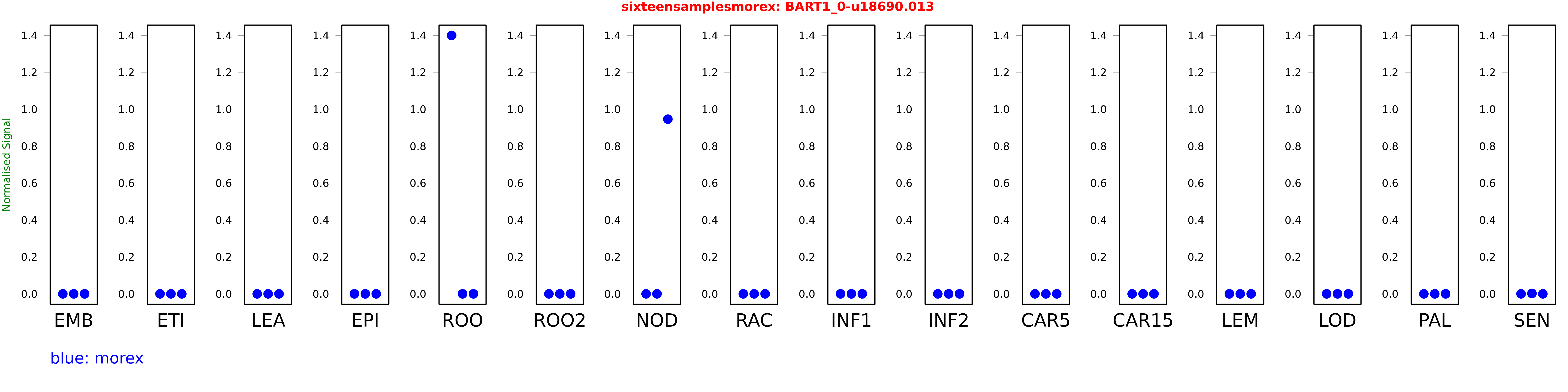
| Sample | Treatment | Rep 1 | Rep 2 | Rep 3 |
|---|---|---|---|---|
| morex | EMB | 0 | 0 | 0.000000938766 |
| morex | ETI | 0 | 0 | 0 |
| morex | LEA | 0 | 0 | 0 |
| morex | EPI | 0 | 0 | 0 |
| morex | ROO | 1.39972 | 0 | 0 |
| morex | ROO2 | 0 | 0 | 0 |
| morex | NOD | 0 | 0 | 0.946055 |
| morex | RAC | 0 | 0 | 0 |
| morex | INF1 | 0 | 0 | 0 |
| morex | INF2 | 0 | 0 | 0 |
| morex | CAR5 | 0 | 0 | 0 |
| morex | CAR15 | 0 | 0 | 0 |
| morex | LEM | 0 | 0 | 0 |
| morex | LOD | 0 | 0.0000749724 | 0 |
| morex | PAL | 0 | 0.0000168146 | 0 |
| morex | SEN | 0 | 0.00217729 | 0 |
Homology to Model Species (BLASTX to E-value < 1e-30)
| Database | Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Rice PP7 | LOC_Os01g08700.8 | +3 | 0.0 | 851 | 435/514 (85%) | protein|GIGANTEA, putative, expressed |
| TAIR PP10 | AT1G22770 @ ARAPORT AT1G22770.1 @ TAIR |
+3 | 1e-177 | 556 | 309/530 (58%) | Symbols: GI, FB | gigantea protein (GI) | chr1:8062398-8067447 FORWARD LENGTH=1173 |
| BRACH PP3 | Bradi2g05226.1.p | +3 | 0.0 | 893 | 457/514 (89%) | pacid=32776210 transcript=Bradi2g05226.1 locus=Bradi2g05226 ID=Bradi2g05226.1.v3.1 annot-version=v3.1 |
Barley PseudoMolecules GBrowse
Click here to see more tracks within GBrowse -->
grey: non-coding, green: Barley Morex IBSC 2017 Transcript CDS, red: Barley RTD exons
CDS Sequence (3726 bp)
>BART1_0-u18690.013 3726 150831_barley_pseudomolecules CCCAAATTTGGTTAGCAGTCATCTGACTTGCAGTTATTGCCCCTCTGAAATATGTGGATT GTGTATTATTATGTTCAGATTTGTATGAACTTGTACAGTAGTTATAAGCTTACAACGTAG CTCTGCTATTGTTTTTTCTTTCTCCTTTTTCTTCATTCTGGTTTCTGTTTCTGGGGTCCC AGTTCCGAGTTGTTTTCTTTGTTTCTGCGTTGTGCCATTTTGGTGTTACTCATCTAATAC AAACGGCACATCTTTTGGTCTGATGTGTCATGTGTGTTCGAGAAAGACGGCATATATCTT TTATTTCTTTATTGAGAATGGTTGCTCCTTTTCTAGATGAGAATTTGATGCTTCTAGTTG TGTAAAAATACATCAATTTTACACACATAAGCTCTTTTTAAAGTCCAAAAGACAATCCTC AGCTCCGAAACAAAACAGTTCAATTTGCAACCTAAAACGACCCCCCCCCGCCGCGCGCGC GCCAAAAGAAAGAACATCTTTTGGGAGACGTGCCATTTACGTCCAAGGCAGCCATCTCTC CAACTCGAGGAATCATGTTGAGCAGAATTGGGTCTGGGACCAATGATGAAGATGAGTAGA TGACTGAGCCCTTGGATCCTGATCCAAGGGTCAGAGTTGGCGATTAGATGTGTTCCTATA CTATCTTATTTCCCCAAGCATCTGGAATCATTCCATTCCATCCCATTCTCAAATCTCAAA GTCCAGGATGGTTCCATTCCAGTCTACTTCATTCCTCAAACCACTAACGGTTTTAGAGTT GAAATCCTTTTTCTGACTCTGTTAAGGGCTGGGAATGTAAACGAGACTTCTTTTTAGAAA TATGTTGCCAAGGCTATGCAAAATTTGTTCCATTGGCTGGTTGACCTCCTCTCTTGTCAA TTTATGGCAATATTTCTTGTTTTAAAATTTATTAAAATGACATCTATAATAATGTCCCTT TCCTTCTTTCGACTTTCCTTTTGCTATATTTTGCAATGCAGAGACGTTAAGCGCCTATGG TTGGAACTGGTTGATATGATATCTGGTCACATTGTGGGATGCAATCCCCCCTTAGTGGGG GGTGATTGAAGAAGTCAACAATAGGAGAACTTGCATCTTGCTGAAGTTTTCTGGTACTGA GGCTCTTAAACTACCAAGTATGTCAGCGTCAAATGGGAAGTGGATTGATGGACTCCAGTT CTCATCACTGTTCTGGCCCCCGCCACACGATGCGCAGCAGAAACAGGCACAAATTTTAGC CTATGTTGAGTACTTTGGTCAGTTCACATCTGACAGCGAGCAATTCCCGGAGGATGTAGC TCAGCTAATTCAAACTTGCTATCCATCAAAAGAAAAGCGGTTGGTAGATGAAGTATTAGC AACTTTTGTTCTCCATCACCCCGAGCATGGTCATGCAGTTGTACATCCAATTCTTTCACG CATCATAGACGGGACACTGAGTTATGATAGTCATGGTTCCCCATTCAATTCCTTCATCTC CTTATTTACCCAAAGTTCTGAGAAAGAGTACTCAGAGCAGTGGGCCTTGGCCTGTGGAGA GATTCTTAGAGTTCTAACTCACTACAACAGGCCAATCTTTAAAGTTGCAGACTGTAACAA CACCTCTGACCAGGCTACAACAAGTTGTTCTGCACAGGAGAAAGCTAATTACTCTCCAGG AAATGAACCTGAACGGAAGCCATTGAGGCCATTATCTCCTTGGATCACAGATATTTTGCT AACTGCACCTTTAGGCATTAGAAGCGACTATTTTAGATGGTGTGGTGGAGTTATGGGAAA ATACGCAGCTGGTGGAGAATTGAAGCCTCCAACAACTGCTTACAGCCGAGGAGCTGGTAA GCACCCACAACTCATGCCATCCACCCCAAGATGGGCTGTTGCCAATGGAGCTGGAGTTAT TTTAAGTGTCTGTGATGAGGAAGTAGCTCGTTATGAGACAGCAAACTTAACCGCGGCAGC TGTTCCCGCCCTTCTGCTACCTCCACCGACAACACCCTTGGATGAGCATTTGGTGGCAGG GCTACCCCCTCTTGAGCCATACGCTCGCTTGTTTCATAGATACTATGCAATTGCCACACC AAGTGCTACACAAAGATTGCTTTTTGGTCTTCTTGAAGCACCACCATCATGGGCTCCAGA TGCACTTGATGCAGCAGTTCAGCTTGTTGAACTCCTTCGGGCAGCTGAAGATTATGCTAC TGGCATGCGGGCATGCTGAAGGAGTTGAAGTGCAACATGAACCACTGGGTGGCTATGTAT CATCATACAAAAGACAGCTGGAAGTTCCTGCATCTGAAACCACAATTGATGCCACTGCAC AAGGCATTGCTTCCTTGCTGTGTGCTCATGGTCCTGATGTTGAGTGGAGAATATGTACCA TCTGGGAAGCTGCCTATGGTTTGTTACCTCTGAATTCATCAGCAGTTGATTTGCCCGAAA TCGTTGTAGCTGCTCCGCTTCAGCCACCTACTTTGTCATGGAGCCTATACTTGCCACTGT TGAAAGTATTCGAGTATCTACCTCGTGGAAGTCCATCTGAAGCATGCCTTATGAGAATAT TTGTGGCAACAGTTGAAGCTATACTCAGAAGAACTTTCCCTTCGGAAACCTCTGAATCAT CTAAAAGACCAAGAAGTCAATCCAAGAACCTTGCTGTTGCTGAACTCCGTACAATGATAC ATTCACTCTTTGTTGAATCATGTGCTTCAATGAACCTTGCTTCCCGGTTGTTGTTTGTTG TATTAACTGTTTGCGTCAGTCATCAAGCTTTGCCAGGGGGCAGCAAAAGACCAACGGGTA GTGAAAACCATTCTTCTGAGGAGGCCACTGAGGACCCAAGATTAACCAATGGAAGAAATA AGGTCAAGAAGAAACAAGGGCCTGTTGGTACATTTGACTCGTATGTGCTGGCTGCTGTTT GTGCCTTATCTTGTGAGCTTCAGCTGTTCCCTATCCTTTGCAAGAGTGCAACAAACTCAA AAGTAAAAGACTCTATAAAGATCCTGAAGCCTGGAAAAAACAATGGGATCAGTAATGAGC TACAGAATAGCATTAGCTCAGCAATTCTCCATACTCGTAGAATTCTTGGCATCCTGGAAG CTCTTTTCTCCTTGAAGCCATCATCAGTTGGTACCTCCTGGAACTATAGTTCAAATGAGA TAGTTGCAGCGGCTATGGTTGCCGCTCATGTTTCTGAGTTATTTCGCCGGTCGAGGCCAT GCCTAAATGCACTATCTTCACTGAAGCGATGTAAGTGGGATGCTGAGATTTCTACCAGGG CATCATCCCTTTACCATTTGATCGATTTGCATGGTAAAACTGTGTCCTCCATCGTGAACA AAGCTGAGCCTCTAGAAGCTCACCTGACTTTTACATCAGTAAAGAGAGATGGTCAACAAC ACATTGAGGAAAACAGCACCAGCTCATCGGGTAATGGCAACTTGGAAAAGAAGAATGCTT CAGCCTCACACATGAAAAATGGTTTTTCAAGACCACTCTTGAAATGCTCAGAAGAGGCTA GGCGAAATGGTAATGTTGCAAGTACATCCGGGAAAGTTCCTGCAACTTTACAGGCTGAAG CATCTGATTTGGCTAACTTCCTTACCATGGATAGAAATGGGGGTTATCGAGGCTCTCAGA CTCTCCTAAGTTCTGTTATCTCAGAAAAACAGGAATTATGCTTCTCTGTTGTCTCATTGC TCTGGC
Protein Sequence (568 aa)
>BART1_0-u18690.013 568 150831_barley_pseudomolecules MSIWWQGYPLLSHTLACFIDTMQLPHQVLHKDCFLVFLKHHHHGLQMHLMQQFSLLNSFG QLKIMLLACGHAEGVEVQHEPLGGYVSSYKRQLEVPASETTIDATAQGIASLLCAHGPDV EWRICTIWEAAYGLLPLNSSAVDLPEIVVAAPLQPPTLSWSLYLPLLKVFEYLPRGSPSE ACLMRIFVATVEAILRRTFPSETSESSKRPRSQSKNLAVAELRTMIHSLFVESCASMNLA SRLLFVVLTVCVSHQALPGGSKRPTGSENHSSEEATEDPRLTNGRNKVKKKQGPVGTFDS YVLAAVCALSCELQLFPILCKSATNSKVKDSIKILKPGKNNGISNELQNSISSAILHTRR ILGILEALFSLKPSSVGTSWNYSSNEIVAAAMVAAHVSELFRRSRPCLNALSSLKRCKWD AEISTRASSLYHLIDLHGKTVSSIVNKAEPLEAHLTFTSVKRDGQQHIEENSTSSSGNGN LEKKNASASHMKNGFSRPLLKCSEEARRNGNVASTSGKVPATLQAEASDLANFLTMDRNG GYRGSQTLLSSVISEKQELCFSVVSLLW
