- Transcript BART1_0-u18690.001
- Transcript IDBART1_0-u18690.001
- Gene IDBART1_0-u18690
Exon Structure of BART1_0-u18690.001
| Chromosome | Exon | Start | Stop | Direction |
|---|---|---|---|---|
| chr3H | 1 | 67423171 | 67421725 | - |
| chr3H | 2 | 67425398 | 67425189 | - |
| chr3H | 3 | 67425680 | 67425530 | - |
| chr3H | 4 | 67425991 | 67425751 | - |
| chr3H | 5 | 67426259 | 67426201 | - |
| chr3H | 6 | 67426950 | 67426694 | - |
| chr3H | 7 | 67427189 | 67427047 | - |
| chr3H | 8 | 67427332 | 67427278 | - |
| chr3H | 9 | 67427506 | 67427429 | - |
| chr3H | 10 | 67427717 | 67427601 | - |
| chr3H | 11 | 67430167 | 67429767 | - |
grey: non-coding, green: Barley Morex IBSC 2017 Transcript CDS, red: Barley RTD exons
Salmon TPM Values: sixteensamplesmorex
These TPM values were generated by using the RNA-seq data from a 16-tissue experiment in Morex (published here) were calculated using Salmon (version Salmon-0.8.2) using BaRTv1.0-QUASI as the reference transcript dataset. Click here for more information about the RNA-seq experiment and materials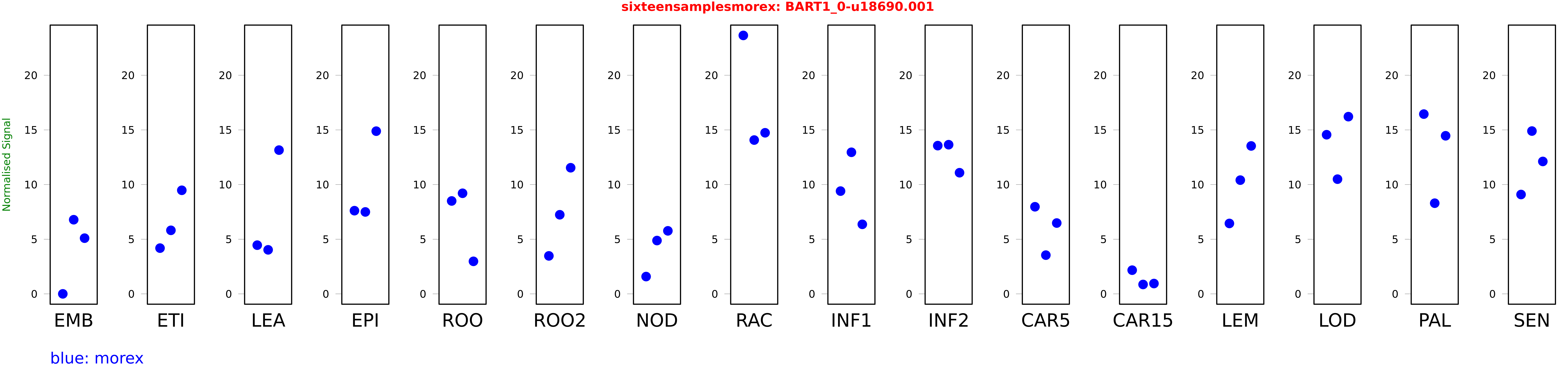
| Sample | Treatment | Rep 1 | Rep 2 | Rep 3 |
|---|---|---|---|---|
| morex | EMB | 0 | 6.78275 | 5.09532 |
| morex | ETI | 4.18434 | 5.8132 | 9.47612 |
| morex | LEA | 4.45519 | 4.02733 | 13.1525 |
| morex | EPI | 7.61035 | 7.49557 | 14.8858 |
| morex | ROO | 8.49947 | 9.20318 | 2.97596 |
| morex | ROO2 | 3.47375 | 7.23724 | 11.5387 |
| morex | NOD | 1.58163 | 4.87729 | 5.76553 |
| morex | RAC | 23.6455 | 14.0722 | 14.7374 |
| morex | INF1 | 9.40176 | 12.9536 | 6.36082 |
| morex | INF2 | 13.5621 | 13.6476 | 11.0834 |
| morex | CAR5 | 7.96822 | 3.54257 | 6.47979 |
| morex | CAR15 | 2.16664 | 0.859028 | 0.944008 |
| morex | LEM | 6.4422 | 10.4055 | 13.5333 |
| morex | LOD | 14.5598 | 10.4943 | 16.2137 |
| morex | PAL | 16.4475 | 8.29445 | 14.4633 |
| morex | SEN | 9.08975 | 14.8957 | 12.1152 |
Homology to Model Species (BLASTX to E-value < 1e-30)
| Database | Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Rice PP7 | LOC_Os01g08700.6 | +3 | 0.0 | 1566 | 807/912 (88%) | protein|GIGANTEA, putative, expressed |
| TAIR PP10 | AT1G22770 @ ARAPORT AT1G22770.1 @ TAIR |
+3 | 0.0 | 1145 | 624/929 (67%) | Symbols: GI, FB | gigantea protein (GI) | chr1:8062398-8067447 FORWARD LENGTH=1173 |
| BRACH PP3 | Bradi2g05226.1.p | +3 | 0.0 | 1643 | 846/906 (93%) | pacid=32776210 transcript=Bradi2g05226.1 locus=Bradi2g05226 ID=Bradi2g05226.1.v3.1 annot-version=v3.1 |
Barley PseudoMolecules GBrowse
Click here to see more tracks within GBrowse -->
grey: non-coding, green: Barley Morex IBSC 2017 Transcript CDS, red: Barley RTD exons
CDS Sequence (3159 bp)
>BART1_0-u18690.001 3159 150831_barley_pseudomolecules CCGAAAATATCAAGGCCATGTCAGCCGCGCCCCCTGGCGATCCCGCCAGCCCCCAGCCGC GCACGCGCGAATAAGTATCTCCGCCCACCCACCGGTCGCCGACCCGCGGGCCCGCCGCCC CCACAGCCCAGGTGTCAGTCACGCAGCGCACCGCGCGGGCTCCACCCCTCGCCCCGCGAG GCGAGTGAAATAACGGAGCCAACGCTACACAGACCACCGCCTACCCCTCCCATTACGGAA AAAATCTTTTTCTCGCCTCTCCTCGCCCCTCACATTTTTCTCTCTCCTCCTCCTCCTCCT CTCTGCCTTCGCCTCTGCCTCAGCCTCGCCCGCCGCGCAATCCCGTCGCCGGCGCCGCCG ATTCGCCGCCCGAGCTCCGTATTGGGGCGCTCCCGACCGAGTTTCTTGAGCAGGCTCTTA AACTACCAAGTATGTCAGCGTCAAATGGGAAGTGGATTGATGGACTCCAGTTCTCATCAC TGTTCTGGCCCCCGCCACACGATGCGCAGCAGAAACAGGCACAAATTTTAGCCTATGTTG AGTACTTTGGTCAGTTCACATCTGACAGCGAGCAATTCCCGGAGGATGTAGCTCAGCTAA TTCAAACTTGCTATCCATCAAAAGAAAAGCGGTTGGTAGATGAAGTATTAGCAACTTTTG TTCTCCATCACCCCGAGCATGGTCATGCAGTTGTACATCCAATTCTTTCACGCATCATAG ACGGGACACTGAGTTATGATAGTCATGGTTCCCCATTCAATTCCTTCATCTCCTTATTTA CCCAAAGTTCTGAGAAAGAGTACTCAGAGCAGTGGGCCTTGGCCTGTGGAGAGATTCTTA GAGTTCTAACTCACTACAACAGGCCAATCTTTAAAGTTGCAGACTGTAACAACACCTCTG ACCAGGCTACAACAAGTTGTTCTGCACAGGAGAAAGCTAATTACTCTCCAGGAAATGAAC CTGAACGGAAGCCATTGAGGCCATTATCTCCTTGGATCACAGATATTTTGCTAACTGCAC CTTTAGGCATTAGAAGCGACTATTTTAGATGGTGTGGTGGAGTTATGGGAAAATACGCAG CTGGTGGAGAATTGAAGCCTCCAACAACTGCTTACAGCCGAGGAGCTGGTAAGCACCCAC AACTCATGCCATCCACCCCAAGATGGGCTGTTGCCAATGGAGCTGGAGTTATTTTAAGTG TCTGTGATGAGGAAGTAGCTCGTTATGAGACAGCAAACTTAACCGCGGCAGCTGTTCCCG CCCTTCTGCTACCTCCACCGACAACACCCTTGGATGAGCATTTGGTGGCAGGGCTACCCC CTCTTGAGCCATACGCTCGCTTGTTTCATAGATACTATGCAATTGCCACACCAAGTGCTA CACAAAGATTGCTTTTTGGTCTTCTTGAAGCACCACCATCATGGGCTCCAGATGCACTTG ATGCAGCAGTTCAGCTTGTTGAACTCCTTCGGGCAGCTGAAGATTATGCTACTGGCATGC GGCTTCCAAAAAATTGGTTGCATCTTCATTTCTTGCGTGCGATTGGAACTGCAATGTCTA TGAGGGCTGGTATTGCTGCCGATACAGCTGCTGCGTTGCTTTTTCGCATACTATCCCAAC CAACGTTGCTTTTTCCTCCACTAAGGCATGCTGAAGGAGTTGAAGTGCAACATGAACCAC TGGGTGGCTATGTATCATCATACAAAAGACAGCTGGAAGTTCCTGCATCTGAAACCACAA TTGATGCCACTGCACAAGGCATTGCTTCCTTGCTGTGTGCTCATGGTCCTGATGTTGAGT GGAGAATATGTACCATCTGGGAAGCTGCCTATGGTTTGTTACCTCTGAATTCATCAGCAG TTGATTTGCCCGAAATCGTTGTAGCTGCTCCGCTTCAGCCACCTACTTTGTCATGGAGCC TATACTTGCCACTGTTGAAAGTATTCGAGTATCTACCTCGTGGAAGTCCATCTGAAGCAT GCCTTATGAGAATATTTGTGGCAACAGTTGAAGCTATACTCAGAAGAACTTTCCCTTCGG AAACCTCTGAATCATCTAAAAGACCAAGAAGTCAATCCAAGAACCTTGCTGTTGCTGAAC TCCGTACAATGATACATTCACTCTTTGTTGAATCATGTGCTTCAATGAACCTTGCTTCCC GGTTGTTGTTTGTTGTATTAACTGTTTGCGTCAGTCATCAAGCTTTGCCAGGGGGCAGCA AAAGACCAACGGGTAGTGAAAACCATTCTTCTGAGGAGGCCACTGAGGACCCAAGATTAA CCAATGGAAGAAATAAGGTCAAGAAGAAACAAGGGCCTGTTGGTACATTTGACTCGTATG TGCTGGCTGCTGTTTGTGCCTTATCTTGTGAGCTTCAGCTGTTCCCTATCCTTTGCAAGA GTGCAACAAACTCAAAAGTAAAAGACTCTATAAAGATCCTGAAGCCTGGAAAAAACAATG GGATCAGTAATGAGCTACAGAATAGCATTAGCTCAGCAATTCTCCATACTCGTAGAATTC TTGGCATCCTGGAAGCTCTTTTCTCCTTGAAGCCATCATCAGTTGGTACCTCCTGGAACT ATAGTTCAAATGAGATAGTTGCAGCGGCTATGGTTGCCGCTCATGTTTCTGAGTTATTTC GCCGGTCGAGGCCATGCCTAAATGCACTATCTTCACTGAAGCGATGTAAGTGGGATGCTG AGATTTCTACCAGGGCATCATCCCTTTACCATTTGATCGATTTGCATGGTAAAACTGTGT CCTCCATCGTGAACAAAGCTGAGCCTCTAGAAGCTCACCTGACTTTTACATCAGTAAAGA GAGATGGTCAACAACACATTGAGGAAAACAGCACCAGCTCATCGGGTAATGGCAACTTGG AAAAGAAGAATGCTTCAGCCTCACACATGAAAAATGGTTTTTCAAGACCACTCTTGAAAT GCTCAGAAGAGGCTAGGCGAAATGGTAATGTTGCAAGTACATCCGGGAAAGTTCCTGCAA CTTTACAGGCTGAAGCATCTGATTTGGCTAACTTCCTTACCATGGATAGAAATGGGGGTT ATCGAGGCTCTCAGACTCTCCTAAGTTCTGTTATCTCAGAAAAACAGGAATTATGCTTCT CTGTTGTCTCATTGCTCTGGCATAAGCTTGCATGCNNNN
Protein Sequence (909 aa)
>BART1_0-u18690.001 909 150831_barley_pseudomolecules MSASNGKWIDGLQFSSLFWPPPHDAQQKQAQILAYVEYFGQFTSDSEQFPEDVAQLIQTC YPSKEKRLVDEVLATFVLHHPEHGHAVVHPILSRIIDGTLSYDSHGSPFNSFISLFTQSS EKEYSEQWALACGEILRVLTHYNRPIFKVADCNNTSDQATTSCSAQEKANYSPGNEPERK PLRPLSPWITDILLTAPLGIRSDYFRWCGGVMGKYAAGGELKPPTTAYSRGAGKHPQLMP STPRWAVANGAGVILSVCDEEVARYETANLTAAAVPALLLPPPTTPLDEHLVAGLPPLEP YARLFHRYYAIATPSATQRLLFGLLEAPPSWAPDALDAAVQLVELLRAAEDYATGMRLPK NWLHLHFLRAIGTAMSMRAGIAADTAAALLFRILSQPTLLFPPLRHAEGVEVQHEPLGGY VSSYKRQLEVPASETTIDATAQGIASLLCAHGPDVEWRICTIWEAAYGLLPLNSSAVDLP EIVVAAPLQPPTLSWSLYLPLLKVFEYLPRGSPSEACLMRIFVATVEAILRRTFPSETSE SSKRPRSQSKNLAVAELRTMIHSLFVESCASMNLASRLLFVVLTVCVSHQALPGGSKRPT GSENHSSEEATEDPRLTNGRNKVKKKQGPVGTFDSYVLAAVCALSCELQLFPILCKSATN SKVKDSIKILKPGKNNGISNELQNSISSAILHTRRILGILEALFSLKPSSVGTSWNYSSN EIVAAAMVAAHVSELFRRSRPCLNALSSLKRCKWDAEISTRASSLYHLIDLHGKTVSSIV NKAEPLEAHLTFTSVKRDGQQHIEENSTSSSGNGNLEKKNASASHMKNGFSRPLLKCSEE ARRNGNVASTSGKVPATLQAEASDLANFLTMDRNGGYRGSQTLLSSVISEKQELCFSVVS LLWHKLACX
