- Transcript BART1_0-u18118.002
- Transcript IDBART1_0-u18118.002
- Gene IDBART1_0-u18118
Exon Structure of BART1_0-u18118.002
| Chromosome | Exon | Start | Stop | Direction |
|---|---|---|---|---|
| chr3H | 1 | 30560371 | 30559687 | + |
| chr3H | 2 | 30560762 | 30560701 | + |
| chr3H | 3 | 30560965 | 30560882 | + |
| chr3H | 4 | 30561371 | 30561078 | + |
| chr3H | 5 | 30561823 | 30561684 | + |
| chr3H | 6 | 30562313 | 30562226 | + |
| chr3H | 7 | 30562615 | 30562424 | + |
| chr3H | 8 | 30562874 | 30562713 | + |
| chr3H | 9 | 30563162 | 30562969 | + |
| chr3H | 10 | 30563897 | 30563747 | + |
| chr3H | 11 | 30565839 | 30564019 | + |
grey: non-coding, green: Barley Morex IBSC 2017 Transcript CDS, red: Barley RTD exons
Salmon TPM Values: sixteensamplesmorex
These TPM values were generated by using the RNA-seq data from a 16-tissue experiment in Morex (published here) were calculated using Salmon (version Salmon-0.8.2) using BaRTv1.0-QUASI as the reference transcript dataset. Click here for more information about the RNA-seq experiment and materials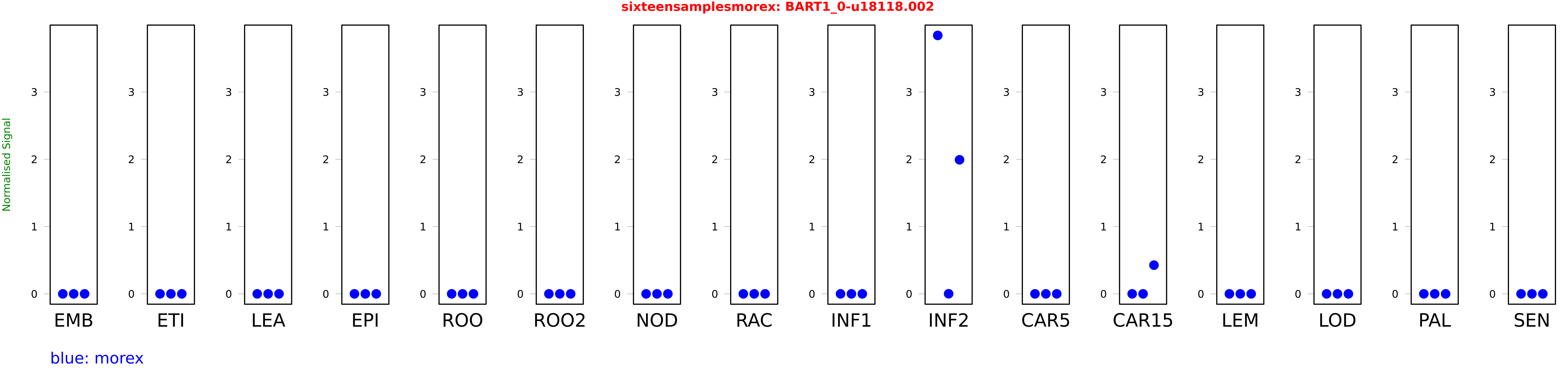
| Sample | Treatment | Rep 1 | Rep 2 | Rep 3 |
|---|---|---|---|---|
| morex | EMB | 0 | 0 | 0 |
| morex | ETI | 0 | 0 | 0 |
| morex | LEA | 0 | 0.000000291152 | 0.000257504 |
| morex | EPI | 0 | 0 | 0 |
| morex | ROO | 0 | 0 | 0 |
| morex | ROO2 | 0.000000385562 | 0 | 0 |
| morex | NOD | 0 | 0 | 0 |
| morex | RAC | 0.00000000692752 | 0.0000000380012 | 0.0000139237 |
| morex | INF1 | 0 | 0 | 0 |
| morex | INF2 | 3.83941 | 0.00136525 | 1.99172 |
| morex | CAR5 | 0 | 0 | 0 |
| morex | CAR15 | 0 | 0 | 0.426505 |
| morex | LEM | 0 | 0 | 0.00000000367509 |
| morex | LOD | 0 | 0 | 0 |
| morex | PAL | 0 | 0 | 0 |
| morex | SEN | 0 | 0 | 0 |
Homology to Model Species (BLASTX to E-value < 1e-30)
| Database | Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Rice PP7 | LOC_Os11g41910.1 | +1 | 0.0 | 564 | 277/317 (87%) | protein|GTP-binding protein, putative, expressed |
| TAIR PP10 | AT3G12080 @ ARAPORT AT3G12080.1 @ TAIR |
+1 | 0.0 | 748 | 370/497 (74%) | Symbols: emb2738 | GTP-binding family protein | chr3:3847851-3851980 FORWARD LENGTH=663 |
| BRACH PP3 | Bradi4g12180.1.p | +1 | 0.0 | 950 | 494/528 (94%) | pacid=32786407 transcript=Bradi4g12180.1 locus=Bradi4g12180 ID=Bradi4g12180.1.v3.1 annot-version=v3.1 |
Barley PseudoMolecules GBrowse
Click here to see more tracks within GBrowse -->
grey: non-coding, green: Barley Morex IBSC 2017 Transcript CDS, red: Barley RTD exons
CDS Sequence (3873 bp)
>BART1_0-u18118.002 3873 150831_barley_pseudomolecules GACGTACTGAAAAGAACATTCAAGGGGGATATGCCGCAATAGTAGTAATTTATTAAAGAC CGTAACGATGCGTTATTGTTCATTCACATGTAGCGACTTACCCTGGTGGCTAGCAGCAGT GTTGGAAAAGGTACGTCAAGTTTTCTATTCCTCGCTGCTGCCCTACGGGCTGCCTTTCGG TGTGCCGGCCCAGCCGCGCGCCTTGTGCGTCGCGCTGGTTATTCGCCGCAAAATGCGGTA AATAGGACAGGAGGTCCCCGCCGGCTGTAGATGCTCTTATCCCCTTCCGTCCCCCGCTGC CCCAGGCCTCGCGCTCTACGGCGCGCCATGGCCATGCCTTCCACTCCTTCCTCCTCGCGG CCGCCCATGCTGCACCCCGCCTCCAAAACTCCGAACCCGCCCCGCCTGCTGCCGCTCGTC ACCAGAGGCGCTGCTGCCCCTCGCCCTCCTCTGCTCATCTCCTTCGCTCCGCCCGCGCCT GGTCGTCGCGGGCTCCGCGCCGCTGCCGCCCAACAACCCGCCTACGAAGATGGGGAGGAG GAAGAAGATGGGGAGGAGGATGGGGAGTGTTACAGCGAGGAGGAGGAGGAGGATGAGGAG GAGTTGGACGTGGAGGCGATGGAGGAAGAGGCGCGGCGCGCCGCCGCCGACCTCGCCGCG CGCCTCGACCGCGAGCTCCGCGTCGATGGTGATGTTCGGGAGAAAAGAAGAACCATGAGG GACAAGACATCAACAACATCTAAACATATCCCAGACAGTAAGCTTCCAAAGGTGGCTATT ATTGGTAGACCTAATGTGGGTAAATCTGCGCTGTTCAATCGGCTTGTCGGGGGTAACAGG GCTATCGTTGTTGATGAACCTGGTGTAACAAGGGATCGTTTGTATGGACGATCTTATTGG GGTGATCAAGAGTTTATGGTTATTGATACTGGGGGTGTCATTACTCTATCGAAGTCTCAA GCAGGTGTAATGGAAGAACTTGCCATCACAACTACTGTTGGTATGGATGGGATTCCAATG GCCTCTCGAGAAGCTGCTATTGCTAGGATGCCATCAATGATTGAGAAGCAAGCTGTTGCC GCCGTTGAAGAAGCAGCTGTCCTTCTTTTCCTTGTGGATGGTCAGGCTGGTCTTGTGGCA GCTGATATAGAAATTGCTGATTGGTTACGTCGCAACTTCTCACACAAGTGCATCATACTT GCTGTAAACAAATGTGAATCGCCACGGAAAGGGCAAATGCAAGCATTAGAATTTTGGTCA TTAGGGTTTTCACCTTTACCAATATCTGCTATTACCGGCACTGGAACTGGAGAGCTCCTG GATATGGTCTCTTCGGAACTGAAAAAGTTCGAGGGACTAGAAGGATTGGATGATGTTGAA GAAGACGAAAACCGCGTTCCTGCTATTTCTATTGTTGGAAGACCAAATGTTGGAAAAAGT AGCATTCTAAATGCTTTGGTGGGAGAAGATAGAACTATTGTGAGCCCAGTTAGTGGTACC ACCCGTGATGCCATTGATACTGAGTTAACCACAGAGGATGGGCAGAAATACAAACTCATC GATACGGCTGGCATCCGACGGAGAGCAGCAGTTGCTTCTGCTGGCAGCACAACTGAAACA CTTTCAGTAAAGCGTGCATTTAGTGCAATTCGTCGATCTGATGTAGTTGCCCTTGTTATT GAAGCGATGGCCTGCGTCTCTGAGCAGGATTATAAAATTGCAGAAAGGATTGAGAAAGAG GGAAAGGCATGTGTCATTGTTGTGAACAAATGGGATACAATCCCAAACAAGAACAATGAG AGTACTACACATTATGAACAGGATGTAAGAGAGAAGCTTCGTGTACTTGACTGGGCACCT ATTGTTTACTGTTCTGCGATAAATGGGAACAGCGTTGAAAAGATTATTTCTGCTGCTTCT TTGGTTGAAAAGGAAAGGTCTAGAAGACTCGGCACCTCTATTCTTAACCAAGTGATTAGA GAAGCTGTAGCATTCAAAGCACCACCCCGAACAAGAGGTGGCAAAAGAGGCCGCGTTTAT TACACAACACAGGCTGCTGTTCGTCCACCAACGTTTGTTCTCTTCGTCAATGACGCAAAA CTCTTCCCTGAGCCGTATCGGCGGTACATGCACAAGCAGCTCCGGTCTGACGCTGGATTC CCGGGCACGCCTATTCGTCTACTGTGGCGTAGCAGGAAGCGGACTGATAGGCAACAGAGG AGATCAAATACGGAGGCTCGTGGTGCACTTGTAGCGGCGAGTTAGCATGCGAAGCAATGT TGCTACCTTCAAAAACACAAGGAAAATGTGGCGAGCTAGGAGTACCACGGTATCCCTTTT CCTCCTTGTTCACGCGCGTTTCCTGGCTGCGAACGTGTGACTATCAATGTGCCAAGAAGT TAAGGTTCATATTCCTCAACTACTGCCAAAGTTGAAGTGCGGTCACTGTACTCTAATAGT ATAATTTTATGGCCTCTCAACTGCTAGAATGTATGGAATTTTATGGCCCCTCTCTCTCTC TCAGATGAACATCAACTTTTCTGTTTTCATGCGATGTTGCATGCATGTATGATTATTGAT GGTTTCTTTTGTCTTCAATCTCATTTTGATGATGTGCTCATTTGCAACACAGATAGGCCT CGGTACGAAAAAAATGGATTGTGCGTTATTGATTAAAATGACAGCATAAATAGAATTTTA AAGTTGTTAAATAGGTAAAATAACATAATATTTAGATTTCATGCATCACTGTAGATTCAA ATTACATTATAAATAAACTTTTAATAAATTAAATAGGTAAACTAACATTATATTTATATT CTACACATTTTTTGAATCAAAGTTCATATATGATAATTTAAAATTGAAGTTATGGTTTAG AAGATATTTTTTTAAAGTATTTGTTAATTATAACTGAAATGCGGATTAATTAACGACATG GATTGGTCATGGTATGAAAAAATTAAATTGTGCGCTATTGGTTAAAATTACAGCATAAAT AGAATTTTAAAATTATTAAATAGGTAAAATAACATCATATTTAGATTGTACGCATCACTA TTGATTAAAATCACATTGTAAACATAATTTTGAAAAACTTAAATAGGTAAACTAACATTA TATTCAGATTCTACACATGTTTCTAATCAAATTTCATATATAATAATTTAAAATTACAGT TACGGTTTAAAATATATAATTTTTAAAGTATTTGTCACTTATAATTGAGCTGCGGATTAA TTAACAACATGGATCGGCCTCGGTACGAGATATTGATTATATGCTATTGATTAAAATTAC AGCATAAATAGAATTTTAAAATTATTAAATAGGTAAAATAATAAATAGAAATTTAAAATT GTTAAATAGGTAAAATAACATCATATTTAGATTGTACACATCACTATTGATTAAAATTAC ATTATAAATAGAATTTTTAAAACATAAATAGGTAAACTAACATTATGTTCAGATTCTACA CATTTTAAAATTGGAGTTGCAGTTTAAAAGATATGATTTTTGAAGTATTTATCACTTATA ATCGAACTGTAGATTAATTAATAGAAATTGTAGGGAATTTTCTAAAAACATATGAAAAAT GATTCGTTTTTTACTTAAGTCTGGGCTGTAGGTTTATTTTATGAAAACTAAAGAAGGTTT TTACAAAAATTATAAAAAACAATTTCTTTTCTCACTTAAATCCGAACTGCGGGTTAATTT CGGAAAAATACAGGAGTATTTTATAAAATGCACGACGGACGATCAGAAGCACTATTTGTT TTATTATTAGGTAATGAAATGGCTCACTGCTACAGCTTTTAAAAGAACTAGATATTTCCA TTTTATGAAAAGTCCAGGCGTACCATGTCTATG
Protein Sequence (664 aa)
>BART1_0-u18118.002 664 150831_barley_pseudomolecules MLLSPSVPRCPRPRALRRAMAMPSTPSSSRPPMLHPASKTPNPPRLLPLVTRGAAAPRPP LLISFAPPAPGRRGLRAAAAQQPAYEDGEEEEDGEEDGECYSEEEEEDEEELDVEAMEEE ARRAAADLAARLDRELRVDGDVREKRRTMRDKTSTTSKHIPDSKLPKVAIIGRPNVGKSA LFNRLVGGNRAIVVDEPGVTRDRLYGRSYWGDQEFMVIDTGGVITLSKSQAGVMEELAIT TTVGMDGIPMASREAAIARMPSMIEKQAVAAVEEAAVLLFLVDGQAGLVAADIEIADWLR RNFSHKCIILAVNKCESPRKGQMQALEFWSLGFSPLPISAITGTGTGELLDMVSSELKKF EGLEGLDDVEEDENRVPAISIVGRPNVGKSSILNALVGEDRTIVSPVSGTTRDAIDTELT TEDGQKYKLIDTAGIRRRAAVASAGSTTETLSVKRAFSAIRRSDVVALVIEAMACVSEQD YKIAERIEKEGKACVIVVNKWDTIPNKNNESTTHYEQDVREKLRVLDWAPIVYCSAINGN SVEKIISAASLVEKERSRRLGTSILNQVIREAVAFKAPPRTRGGKRGRVYYTTQAAVRPP TFVLFVNDAKLFPEPYRRYMHKQLRSDAGFPGTPIRLLWRSRKRTDRQQRRSNTEARGAL VAAS
