- Transcript BART1_0-u13191.013
- Transcript IDBART1_0-u13191.013
- Gene IDBART1_0-u13191
Exon Structure of BART1_0-u13191.013
| Chromosome | Exon | Start | Stop | Direction |
|---|---|---|---|---|
| chr2H | 1 | 614334067 | 614333986 | + |
| chr2H | 2 | 614334855 | 614334784 | + |
| chr2H | 3 | 614335000 | 614334946 | + |
| chr2H | 4 | 614335184 | 614335079 | + |
| chr2H | 5 | 614336189 | 614336085 | + |
| chr2H | 6 | 614337611 | 614337144 | + |
| chr2H | 7 | 614338348 | 614338217 | + |
| chr2H | 8 | 614338544 | 614338458 | + |
| chr2H | 9 | 614338728 | 614338642 | + |
| chr2H | 10 | 614338934 | 614338838 | + |
| chr2H | 11 | 614339597 | 614339480 | + |
| chr2H | 12 | 614348172 | 614348118 | + |
| chr2H | 13 | 614348432 | 614348266 | + |
| chr2H | 14 | 614349221 | 614349034 | + |
| chr2H | 15 | 614350161 | 614350013 | + |
| chr2H | 16 | 614350541 | 614350292 | + |
| chr2H | 17 | 614351269 | 614351011 | + |
| chr2H | 18 | 614351485 | 614351389 | + |
| chr2H | 19 | 614354844 | 614354764 | + |
| chr2H | 20 | 614355033 | 614354977 | + |
| chr2H | 21 | 614355353 | 614355222 | + |
| chr2H | 22 | 614355517 | 614355431 | + |
| chr2H | 23 | 614355844 | 614355736 | + |
| chr2H | 24 | 614356065 | 614355929 | + |
| chr2H | 25 | 614356910 | 614356806 | + |
| chr2H | 26 | 614357771 | 614357683 | + |
| chr2H | 27 | 614357971 | 614357869 | + |
| chr2H | 28 | 614358168 | 614358046 | + |
| chr2H | 29 | 614358861 | 614358722 | + |
| chr2H | 30 | 614359613 | 614359050 | + |
grey: non-coding, green: Barley Morex IBSC 2017 Transcript CDS, red: Barley RTD exons
Salmon TPM Values: sixteensamplesmorex
These TPM values were generated by using the RNA-seq data from a 16-tissue experiment in Morex (published here) were calculated using Salmon (version Salmon-0.8.2) using BaRTv1.0-QUASI as the reference transcript dataset. Click here for more information about the RNA-seq experiment and materials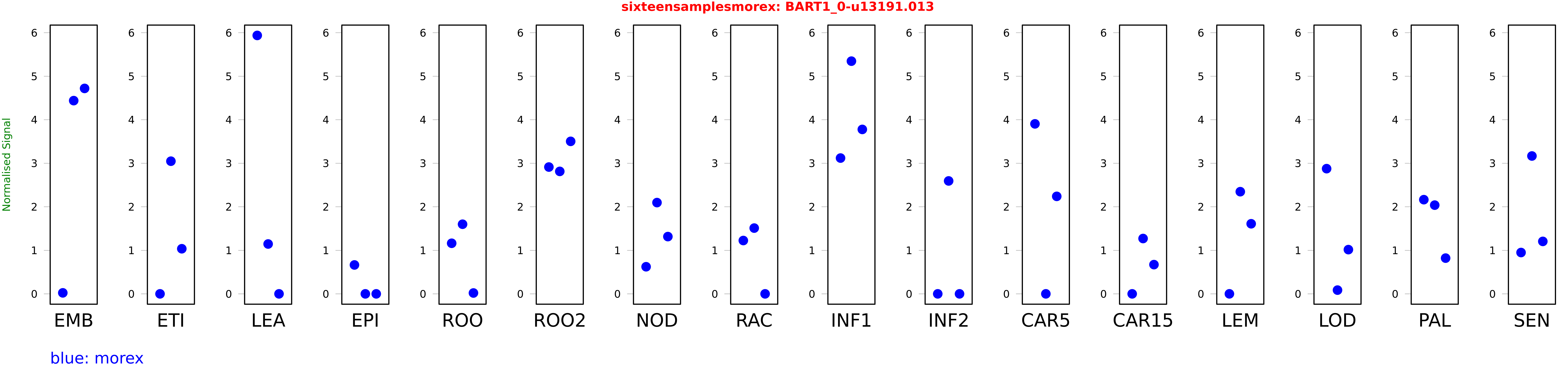
| Sample | Treatment | Rep 1 | Rep 2 | Rep 3 |
|---|---|---|---|---|
| morex | EMB | 0.0228319 | 4.4395 | 4.72011 |
| morex | ETI | 0 | 3.04793 | 1.03628 |
| morex | LEA | 5.93753 | 1.14564 | 0.00000312833 |
| morex | EPI | 0.664077 | 0 | 0.0000000680172 |
| morex | ROO | 1.16218 | 1.59964 | 0.0197935 |
| morex | ROO2 | 2.91354 | 2.8137 | 3.50386 |
| morex | NOD | 0.622812 | 2.09676 | 1.31564 |
| morex | RAC | 1.22541 | 1.51102 | 0 |
| morex | INF1 | 3.11986 | 5.34629 | 3.7781 |
| morex | INF2 | 0 | 2.59447 | 0 |
| morex | CAR5 | 3.90558 | 0.000280075 | 2.24007 |
| morex | CAR15 | 0 | 1.27139 | 0.671603 |
| morex | LEM | 0.000442462 | 2.34744 | 1.61017 |
| morex | LOD | 2.87644 | 0.0859602 | 1.0162 |
| morex | PAL | 2.16402 | 2.03838 | 0.821001 |
| morex | SEN | 0.949579 | 3.16743 | 1.20523 |
Homology to Model Species (BLASTX to E-value < 1e-30)
| Database | Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Rice PP7 | LOC_Os04g41510.1 | +1 | 0.0 | 1271 | 629/786 (80%) | protein|serine/threonine-protein kinase GCN2, putative, expressed |
| TAIR PP10 | AT3G59410 @ ARAPORT AT3G59410.1 @ TAIR |
+1 | 0.0 | 1140 | 611/1190 (51%) | Symbols: GCN2 | protein kinase family protein | chr3:21950575-21959151 FORWARD LENGTH=1241 |
| BRACH PP3 | Bradi5g14307.1.p | +1 | 0.0 | 2126 | 1041/1170 (89%) | pacid=32809296 transcript=Bradi5g14307.1 locus=Bradi5g14307 ID=Bradi5g14307.1.v3.1 annot-version=v3.1 |
Barley PseudoMolecules GBrowse
Click here to see more tracks within GBrowse -->
grey: non-coding, green: Barley Morex IBSC 2017 Transcript CDS, red: Barley RTD exons
CDS Sequence (4301 bp)
>BART1_0-u13191.013 4301 150831_barley_pseudomolecules GCGCTGGGCGCAAGGCGGCCAAGGACCACGCCGCGCAGCTCGAGGGCGACCAGACCGCGC TCACGGACGAGCTCACGGCGCTGGCTGCAATTTTTCTTGAGGACTTCAAAATAACTTCAC AATCACCTAACACCCGATTTAGCATATGTATCAGGCCTTACTCTGACGGCATGGGCTTTG GAGATTTAAATGTTTCGGCTATTCTTGATTTGCTTTGCTGGTTACCCCCACAAGTGCCCA AAGTTGCGAATCATACCCGAGAAAAACTTGTCTAAAGAAGATGCTGATCGGTTACTTTCC CTTCTTTCTGACCAGGCAAACATTTATTCCAGAGAAGGGCGTGTTATGATTTTCGATTTG GTAGAGGCTGCCCAAGAATTCCTTTCAGAAATTGCTCCAGCTGCTGATTCATCAAGTACT GCTCCTCACTTAGGTTCAAGTACAGTACAGGAAACAACTGATGCAGATGTGAAAGCTACC CTTGACAGCAGTCCTTACCCTGGAATCTTTTACATCTACAATTCATTCGATTTGTATAGT CAACTATATGATGATAATAGTTGGCAAAGGCAAGGTTTCGATCCTACAACTGATAATGCC AACAAAAATATTGGATCTCAAGTCAATTCAAATGTTATAAGCAAGAGGAAAACAGTGAAT GAAAAATCCCGTTTTTCAGCTGATAAGGTCAATGCTGCAAAAAGTTCATCTCAGGATAAT TCTGAACAGCAGCATGCCATGAAGCTGGGTGTTGTACGGGAGGTAGTTCCAAGTTTACCT GTTGTTGCAGAGGAAACTGATAATGACAGCAAAACTTTGTCCACAAGCAACGGAGGAGGT ATGGCAGATACTCCAGAGAGGAGTTTCAGCAGTGTACATGAATCCGAGGACTCTGGCCTT GCAGATGAAGGTTGGAATGATGAAGATTCTGCTCCAGACTCTGGCTCTTCAAATGCACCA TCTCATGTTTCAGACATGTTTGATGATGCTTCGCAAAATAAGAAAAGGGATTTAATTCTG GTACACTTGCTTCGGTTAGCCTGTGCGTCAAAAGATTCTCTTTCAGCTGCTTTGCCAGTA ATATCATTGGAGTTATGCAACATAGGGGTTCTTTCTGAATGGGCTAAGCAATTAATTTCT GAATCTCCTGCTGTTTTTGGGGAAACTTTCGATCATGTTTTTGGGCAACAAATGATATCT TCGGAGTGCTCTCTATTTTGGAGGGCTGACAATTCATCGTCTAGGCCAAACTCCCGTTAT TTGAATGATTTTGAAGAGCTCCGTTCACTTGGTCAGGGGGGCTTTGGCCGTGTGGCATTG TGTAAAAACAAGCTCGATGGACGCCAATATGCTGTAAAGAAGATACGCCTCAAGGATAGA AGTCCTCAAGTGAATGAAAAGATTCTCAGAGAAGTTGCCACACTTTCACGACTGCAGCAT CAGCATGTTGTTCGTTACTACCAGGCATGGGTTGAAACTGAATATGGTCAACATCATGTT GTGAACAGCGGGGGGTCACGCACTGCTGAGAGCTCTATGTATAGTTTCGACGAGATCAGC TTATCAGATGCAGGTGCTGGAAATAAGCAGGAATGCACGTACTTGTACATCCAAATGGAG TATTGTCCTAGAACCCTGCGACAGGACTTTGAGACATATAGTTCATCTTTCAATGTCGAC CATGCATGGCACTTATTTCGACAAATCGTGGAAGGCCTAGCACATGTTCATAGTCAAGGC ATCATACACCGGGACTTGACACCTAGCAACATATTTTTTGATGTGCGCAACGATATCAAA ATTGGAGATTTTGGTCTTGCCAAATTTCTAAAGCTGGAGCAGCTGGATCATGACCAATAT ATTCCTACTGAAGGAATGGGCGTTTCAATGGATGGAACAGGTCAAGTAGGTACCTATTTT TATACTGCACCGGAGGTAGAACAGAAGTGGCCACAGATAAACGAAAAGGTTGACATGTAC AGTGCGGGTGTTATATTCTTTGAGCTTTGGCATCCATTTTCAACAGCAATGGAAAGACAT CTTGTTCTCACTGATCTTAAACAGAAAGGCGAGTCTCCAATATCATGGTCAACACAATTT CCTGGTCAATCAAATCTATTAAGACGCTTGCTGTGTCCAAGCCCGTCCGAACGTCCATCT GCGATTGAGCTTTTGCAAAATGACTTGCCTCCTCGGATGGAGGACGAGTGGTTAAATGAT GTACTTAGAATGATACAGACACCAGAAGACACTTACGTTTATGATCGAGTTATATCTACA ATATTTAATGAAGATCGACTGGTCGCCAAAATGCAATGTCAGCATGAAAGCAGTAAAAAA TCCACTTATAAAAATGATAACAGTGAACTCTTGGATTCCATTATTGAGGTTAGCAAAGAG GTTTTTAAGCGACATTGTGCTAAAAGGTTCCAGATAACACCCTTGCATACATTGGATGGG AAATTTACTGAAAATAGTGGAAAGACAGTGAAGATCCTAACGCAAGGAGGAGAGATGCTA GAACTTTGCTATGAGCTGCGAACACCATTTGTTATGAACGTTGCTGCTAACCAGTTATCA TCATGTAAGCGTTATGAAATATCATGGGTTCACAGAAGAGCAGTTGGCCATTCAACTCCT TATCGTTTTCTTCAGGGTGATTTTGACATCATTGGAGGTTCTTCACCAATAACACAGGCC GAAGTTATCAAGGTAGCTTTGGACCTTGTGAGACGTTTTTACAATTCGAAGGCAATAGTT ATTCGACTGAATCATAGCAAACTCGCTGAAGCAGTTTGTTCCTGGGCAGGAGTTCCTCAG GAACGAAGACAGAATGTTGCTGAGTTTCTATCTTCTACTCTTGTTCAATATTGCCCAAAC AAGGCGGATCGTAAGTCACAGTGGAGTTTCATTCGAGGGCAACTATTACAGGATCTTCGT CTTTCTGAAGAAGTTGTTGAAAAGCTACATAAAGCAGATCAGCGGTTTTGTGGATCTGCA GATCTAGTACTTGCTAGATTAAGAGGGACCTTTTTCTATGATAAATCTGCTTGCAAGGCT CTTGATGATCTCTCCACATTCCTTAAATGTTTGAGGGTATGGTCAGTTGAAGAACATATT GCTATTGATGTACTTATGCCTCCTTCAGACTATTACTACTCAGATATGTTTTTTCAGATC TACTCAAAAGAAGGCAGTCCTGCGTCGAGTTCCCATGAAAAGTTGCTTGCTGTTGGCGGG CGATATGATATGTTGATGGAAAATGCTTGGGATAAGACACATAAAACCAAACCCCCGGGT GCTGTTGGTGTGAGCATTGCACTTGAGAAATTCCTACCTAATAATCCTTCATCTGACGTT GGGTTGCCAAGGATTGAAAATAGCATCAGTGTACTAGTCTGTTCAAAGGGCGGCGGCGGA CTCTTAAACGAACGCATGGAACTTGTTGCAGAACTTTGGGAAGCTAACATAAAGGCCCAG TTTGTTCCTCAGGAAGACCCAAGTCTTCAAGAACAATACGAGTACGCCAGTGATCATGAC ATCAAATGCCTTGTGTTTATCACTGAAGCAGGTCTTGCGCAAACTGATCTTGTGAAGGTT AGACATCTCGATGCAAGAAAGGAGAAAGATGTTGAAAAGGAAGAAATTGTGAAGTTCCTG TCTGAAGCAATATCTTTACAGTTCAAAAATCCTACAATCTGGAGCTGATTGGCAGTCTGC TCCTAAGTGAATTTCAGGTGAAAGCAGTGTTGCCTTGCCAAACATTTTCACCTACACCAA ATTGCAGGGTGACTGGTGGCTTTGTTCAATCTTGGTTATCCTGGAGCATTGGCCAAGATT GATGGCTTGGTGTAATCCTGATTGTCCTGGAGCAGTATTGCGGTGAATGCAAGCTGACGG GTAGCTGTTTTAAGGCAATCTTGGCTGCTATTATGTGGATAGCTAGTTGGTGCAGAGTTA CTTCTGCGATGTGATCATGGAGCTGGAATGCTGCACAACGTTCTTGTTGATAGTGGAACG GCTGTGCTACAGGTTCACCCAGTTATGCCCTTTGCAAAAAAATTTGCTTCAAATTCTTCT ACAAGGCAGTCTTTTGCATAGTTCAAATTGTTTACGATCATGTACAGGCCAATTTTCCTT CTTTGATGCTCGTAGGAAATGCCCAGTTTTGCAACTGGCTTAGCTTGGAGAAGACAAGCC AATGCAGTTTTGCTACAATACCGGTCAAAGTAGAATGTATGTAATCTGTTATCTGTGCAC AGTGCGAAAGAGCATCACTTGATTCAACTCCAGTGTTTCCG
Protein Sequence (1172 aa)
>BART1_0-u13191.013 1172 150831_barley_pseudomolecules MFRLFLICFAGYPHKCPKLRIIPEKNLSKEDADRLLSLLSDQANIYSREGRVMIFDLVEA AQEFLSEIAPAADSSSTAPHLGSSTVQETTDADVKATLDSSPYPGIFYIYNSFDLYSQLY DDNSWQRQGFDPTTDNANKNIGSQVNSNVISKRKTVNEKSRFSADKVNAAKSSSQDNSEQ QHAMKLGVVREVVPSLPVVAEETDNDSKTLSTSNGGGMADTPERSFSSVHESEDSGLADE GWNDEDSAPDSGSSNAPSHVSDMFDDASQNKKRDLILVHLLRLACASKDSLSAALPVISL ELCNIGVLSEWAKQLISESPAVFGETFDHVFGQQMISSECSLFWRADNSSSRPNSRYLND FEELRSLGQGGFGRVALCKNKLDGRQYAVKKIRLKDRSPQVNEKILREVATLSRLQHQHV VRYYQAWVETEYGQHHVVNSGGSRTAESSMYSFDEISLSDAGAGNKQECTYLYIQMEYCP RTLRQDFETYSSSFNVDHAWHLFRQIVEGLAHVHSQGIIHRDLTPSNIFFDVRNDIKIGD FGLAKFLKLEQLDHDQYIPTEGMGVSMDGTGQVGTYFYTAPEVEQKWPQINEKVDMYSAG VIFFELWHPFSTAMERHLVLTDLKQKGESPISWSTQFPGQSNLLRRLLCPSPSERPSAIE LLQNDLPPRMEDEWLNDVLRMIQTPEDTYVYDRVISTIFNEDRLVAKMQCQHESSKKSTY KNDNSELLDSIIEVSKEVFKRHCAKRFQITPLHTLDGKFTENSGKTVKILTQGGEMLELC YELRTPFVMNVAANQLSSCKRYEISWVHRRAVGHSTPYRFLQGDFDIIGGSSPITQAEVI KVALDLVRRFYNSKAIVIRLNHSKLAEAVCSWAGVPQERRQNVAEFLSSTLVQYCPNKAD RKSQWSFIRGQLLQDLRLSEEVVEKLHKADQRFCGSADLVLARLRGTFFYDKSACKALDD LSTFLKCLRVWSVEEHIAIDVLMPPSDYYYSDMFFQIYSKEGSPASSSHEKLLAVGGRYD MLMENAWDKTHKTKPPGAVGVSIALEKFLPNNPSSDVGLPRIENSISVLVCSKGGGGLLN ERMELVAELWEANIKAQFVPQEDPSLQEQYEYASDHDIKCLVFITEAGLAQTDLVKVRHL DARKEKDVEKEEIVKFLSEAISLQFKNPTIWS
