- Transcript BART1_0-u07987.003
- Transcript IDBART1_0-u07987.003
- Gene IDBART1_0-u07987
Exon Structure of BART1_0-u07987.003
| Chromosome | Exon | Start | Stop | Direction |
|---|---|---|---|---|
| chr2H | 1 | 29124217 | 29123713 | + |
| chr2H | 2 | 29124505 | 29124321 | + |
| chr2H | 3 | 29124781 | 29124617 | + |
| chr2H | 4 | 29124992 | 29124859 | + |
| chr2H | 5 | 29125344 | 29125189 | + |
| chr2H | 6 | 29125969 | 29125786 | + |
| chr2H | 7 | 29126484 | 29126067 | + |
| chr2H | 8 | 29127230 | 29126567 | + |
| chr2H | 9 | 29128419 | 29127330 | + |
grey: non-coding, green: Barley Morex IBSC 2017 Transcript CDS, red: Barley RTD exons
Salmon TPM Values: sixteensamplesmorex
These TPM values were generated by using the RNA-seq data from a 16-tissue experiment in Morex (published here) were calculated using Salmon (version Salmon-0.8.2) using BaRTv1.0-QUASI as the reference transcript dataset. Click here for more information about the RNA-seq experiment and materials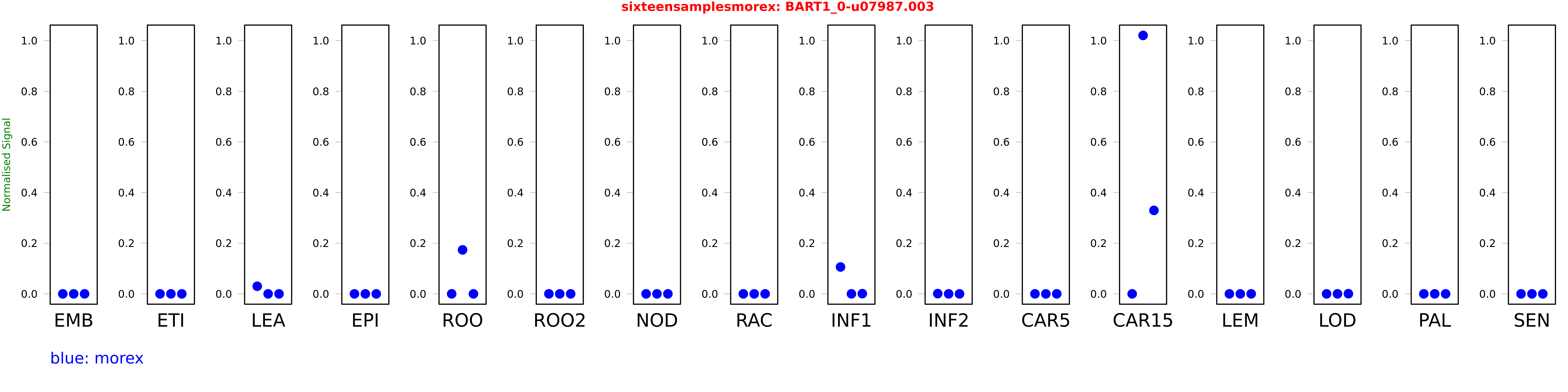
| Sample | Treatment | Rep 1 | Rep 2 | Rep 3 |
|---|---|---|---|---|
| morex | EMB | 0 | 0 | 0 |
| morex | ETI | 0 | 0 | 0 |
| morex | LEA | 0.0300478 | 0 | 0 |
| morex | EPI | 0 | 0 | 0 |
| morex | ROO | 0 | 0.173647 | 0 |
| morex | ROO2 | 0 | 0 | 0 |
| morex | NOD | 0 | 0 | 0 |
| morex | RAC | 0 | 0 | 0 |
| morex | INF1 | 0.106094 | 0.000000012012 | 0.000795837 |
| morex | INF2 | 0.000861726 | 0.00000320106 | 0.00000232683 |
| morex | CAR5 | 0 | 0 | 0 |
| morex | CAR15 | 0.000000148642 | 1.02073 | 0.329712 |
| morex | LEM | 0 | 0.00000000100104 | 0 |
| morex | LOD | 0 | 0 | 0.000636153 |
| morex | PAL | 0 | 0 | 0 |
| morex | SEN | 0 | 0 | 0 |
Homology to Model Species (BLASTX to E-value < 1e-30)
| Database | Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Rice PP7 | LOC_Os07g49460.2 | +3 | 0.0 | 646 | 427/685 (62%) | protein|response regulator receiver domain containing protein, expressed |
| TAIR PP10 | AT5G02810 @ ARAPORT AT5G02810.1 @ TAIR |
+3 | 1e-71 | 254 | 190/454 (42%) | Symbols: PRR7, APRR7 | pseudo-response regulator 7 | chr5:638283-641461 REVERSE LENGTH=727 |
| BRACH PP3 | Bradi1g16490.7.p | +3 | 0.0 | 755 | 475/675 (70%) | pacid=32803660 transcript=Bradi1g16490.7 locus=Bradi1g16490 ID=Bradi1g16490.7.v3.1 annot-version=v3.1 |
Barley PseudoMolecules GBrowse
Click here to see more tracks within GBrowse -->
grey: non-coding, green: Barley Morex IBSC 2017 Transcript CDS, red: Barley RTD exons
CDS Sequence (3501 bp)
>BART1_0-u07987.003 3501 150831_barley_pseudomolecules CCCGATATCTACTCCTCCGCCCCCCTCCCTCGCCTCCCACACCCACCCACACACACAGCA CACCCTCCTCGGCTCGACTCGACTCGCCGCCGCCGCCGCAGTCCATGGCCTCGTCGTCCT CCTCCTGACCGGCAGCAGACCCAAGGTGCGCTCCCGCCCGTCTATGGTCACCCACGCGCT CTGTTTCCGCTCGATTGGGGGATCGAATCACCCGTTTCAATCTCAATCTACGTGCGTGTG GATCCAGGGCACGCTCAGGCTCTCTCTAGCTGCTGCTAATAGTATGTGTCCGGAAATTGC TGGAGATTAACCGGCTCGCACGCCTTGTCTGCTACTGCCCCCCCTTATAGCCGCCGGCAG CTCCATTCCAGGAGACGATTCATTCCGCTGCAGTCTCAACCCCCTTGCTCTTGTGTGTTT TTTTTGCGTTTTGTTTTTATTGGTTTGGTCCGCTGCTCCGCCGCCGTCGACGACGAACGC CGTGGGACTCCCGGATGCGACCGAGGCAGCCTCCTCCTAGCGCCATGGACAATCAGCAGC AGCAGCAGCCGCCGCGCGGGGAGCACGCCGCCCAGCCGCGATGCTGGGAGGAGTTCCTAC ACAGGAAGACCATCCGGGTCCTGCTCGTGGAGACCGACGACTCCACCAGGCAGGTCGTCG CCGCCCTGCTCCGCCACTGCATGTACCAAGTTATCCCCGTCGAGAACGGCCACCAGGCGT GGGCGTATCTTCAGGACATGCAGAGCAACATCGACCTTGTTCTCACAGAGGTGTTCATGC ACGGCGGCCTCTCCGGGATCGACCTGCTCGGCAGGATCATGAACCACGAGGTCTGCAAGG ACATCCCCGTCATCATGATGTCGTCGCACGATTCCATGGGCACCGTCCTCAGCTGCCTCT CAAATGGTGCTGCTGACTTCTTGGCCAAGCCGATACGTAAGAACGAGCTTAAGAACCTTT GGGCGCATGTGTGGCGACGGTCTCACAGCTCCAGTGGCAGTGGTAGTGGAAGTGCCATTC AGACTCAGAAGTGTACCAAATCAAAGAGCGGCGATGATTCCAATAACAACAGCAATGATC GCAACGACGATGCCAGCATGGGGCTCAATGCAAGGGATGGCAGCGATAATGGCAGTGGCA CTCAGAGCTCATGGACAAAGCGTGCCGTTGAGATCGACAGTCCCCAGGACATGTCTCCGG ATCAGTCAGCCGATCCCCCTGAAGGCACTTGTGCCCATGTGAACCATCCCAAGTCAGAGA TATGCAGCAATAGATGGTTACCAGGTACAAATAACAAAAAATGCCAGAAACCAAAAGAAA CCACTAATGGTGATGGATTCAAAGGCAAGGAGTTGGAGATAGGAGCCCCTGGTAATTTGA ACACAGACGATCAATCCTCCCCGAACGAGAGTTCGGTCAAACCAACCGATGGACGGTGTG AGTATCTGCCACAGAACAATTCCAGTGATACGGTCATGGAGAATTTGGAGGAGCCAATTG TTCGAGCTGCTGACTTAATCGGTTCGATGGCCAAAAACATGGACGCCCAACAGGCAGCAG CTAGAGCCATAGATGCTCCTAACTGCTCCTTACAGGCGTCGGAAGGGAAAGACACGGACG GCGAGAACGCTATGCCATATCTTGAGCTGAGCCTGAAGAGGTCGAGATCGACCGGGGAAG GTGCAGGCCCGATCCAGGAGGAACAGAGGAACGTCGTGAGACGGTCAGACCTGTCGGCAT TCACGAGGTACAATATGTGTGCGGTCTCCAATCAAGGCGGGGCAGGGTTCGTCGGGAGCT GCTCACCCAACGGCGACAGCTCCGAGGCCGCGAAAACGGTTGCTGCTCAGATGAAGCAGG GCTCTAACGGCAGCAGCAACAACAATGACATGGGCTCCACCACGAAGAGCGTGGTGACCA AGCCCTGTGGAAATAATAAGGTTTCGCCCATCAACGGCAACACACACACGTCGGCGTTTC ATCGTGTGCAGCCATGGACGCCAGCGACAGCAGCAGGAAAAGACAAGGCTGATGAAGTGA GCAAGAAGAACGTTGCAGCAGCAGCAGCAGCAGCGAAAGAAATGGGTTGTGAAGCACAGA GCAAACACCCCTGTGCAGCGGCCGATGACGTGAATGGTGGATCGGCAGGAGGCACTGCTC AGTCCAGTGTTGTCAATCCTTCGGGTCCGGTTGAAGGCCATACTGCCAACTATGGTAGCA ACTCATTCAGCAATAACAACACCAATAACGGGAGCACCGCAGCTACCACTGCTGCTGCTG CTGCTGTACATGCTGAGACCGGTGGTGGCGTCGACAAAAGAAGTAACATGATGAACATGA AACGGGAGCGCCGTGTGGCCGCCGTGAACAAGTTCAGAGAGAAGAGAAAAGAGAGGAACT TCGGGAAGAAGGTGCGTTACCAGAGCAGGAAGCGGCTGGCCGAGCAGCGCCCACGGGTGC GCTGGCAGTTCGTGCGGCAGCCGCCGGCGCCGGCGACCGTGGAGAGATAACCTCCCGCCA CGCACCTAGCTATACCTAGTACTCTACTAATTAGACAGCTGATTCTTGTGTCTTTGGCCA TGGATCATTGGGAGCTCGTCTCATCTCCTGGAAGAAGAAGAATAACGAGTGACTCCTCCT GCCTGCCTTAGCGACCTACCATAGAACATGCCGGCGGAGGAGGACGAGGGCGATTGTACA TAGTAGGAGCGTGTGGAGCCGATCAGACAGGCTCCCGCTCTTGAATGTAGCAGCAATTGA CCAATCAATGTCTGAGCCTCTAGAATGGTAGTATGAGTGAGTATTGTGCTGTACACTTGT CATCAGCTTGGCCATGGCGGCAGCCTTGTCAGGTGTCATCAATCAACGCATATTTGTGGT TTCTTTCTAGTTACCGGGCAGACATCATCATCGTACTTGTTGCAAATCTGTCTAACCTGT GTCGGCATCACGCGACATGTGGAAGTTTTGTTGCACATTGTCTTTCATTTTAAAAGAAGA AGAAGAAGAAGAAGTAGAAGAAGAAGGTGAAGACCGCTGAGACGAAACCGCATTTGTAGA TTCTTTACTACTACTACGGTGAAGTGCTCCCTGCGTTTCCTATCCAAGCGTTGTAGCATG CACTCGTCTTGCTTCAGCATCCTTGCGCGGCGGACTCCAGCCTTTCGTCGACGAACAAGC ACGAATCTTGGCTGGTGCTGGTGCTGGTGCAGAACACTTGAGAATGTACCGGGGGGACGA AGGAACCGGATCCTGTAGAGGACCACACGGGTAAATCCGTAACCGGTGGACGAGGCGAGT TGCTCCACGATGTTTTAGGTGGTGTTTCTTTTCAAGGGACTAGATTTTTTTTTTAGTCTT AGGGATTAAAGAAAAAAGTCCTTTCAATAGAATCTTTTTCTAGTCTTTTAAGGAAAAGCC CCTCCTGTTTTTTACTTAGGGACTAAAAGTGACTTTTTAGTCTAGTCCCTGGGTAAAGAA ACACCACCTTAACAAGTTGAG
Protein Sequence (671 aa)
>BART1_0-u07987.003 671 150831_barley_pseudomolecules MRPRQPPPSAMDNQQQQQPPRGEHAAQPRCWEEFLHRKTIRVLLVETDDSTRQVVAALLR HCMYQVIPVENGHQAWAYLQDMQSNIDLVLTEVFMHGGLSGIDLLGRIMNHEVCKDIPVI MMSSHDSMGTVLSCLSNGAADFLAKPIRKNELKNLWAHVWRRSHSSSGSGSGSAIQTQKC TKSKSGDDSNNNSNDRNDDASMGLNARDGSDNGSGTQSSWTKRAVEIDSPQDMSPDQSAD PPEGTCAHVNHPKSEICSNRWLPGTNNKKCQKPKETTNGDGFKGKELEIGAPGNLNTDDQ SSPNESSVKPTDGRCEYLPQNNSSDTVMENLEEPIVRAADLIGSMAKNMDAQQAAARAID APNCSLQASEGKDTDGENAMPYLELSLKRSRSTGEGAGPIQEEQRNVVRRSDLSAFTRYN MCAVSNQGGAGFVGSCSPNGDSSEAAKTVAAQMKQGSNGSSNNNDMGSTTKSVVTKPCGN NKVSPINGNTHTSAFHRVQPWTPATAAGKDKADEVSKKNVAAAAAAAKEMGCEAQSKHPC AAADDVNGGSAGGTAQSSVVNPSGPVEGHTANYGSNSFSNNNTNNGSTAATTAAAAAVHA ETGGGVDKRSNMMNMKRERRVAAVNKFREKRKERNFGKKVRYQSRKRLAEQRPRVRWQFV RQPPAPATVER
