Probe CUST_999_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_999_PI426222305 | JHI_St_60k_v1 | DMT400003463 | GTCAACATCACTTACACTACTGAAACCCCGCCTTAAATGTTATTCATTGCTCTGATTTCC |
All Microarray Probes Designed to Gene DMG400001369
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_1048_PI426222305 | JHI_St_60k_v1 | DMT400003464 | ACATGGCTAATGGTACCCTCCGTGATCACTTGTACAATACTGACAATGCTCCTCTTCCAT |
| CUST_999_PI426222305 | JHI_St_60k_v1 | DMT400003463 | GTCAACATCACTTACACTACTGAAACCCCGCCTTAAATGTTATTCATTGCTCTGATTTCC |
| CUST_1316_PI426222305 | JHI_St_60k_v1 | DMT400003465 | CCCTATTTCATAACCCCATTGTTTGACGACGTCAACTCTTTTATGGAATGTCTTATAAAA |
Microarray Signals from CUST_999_PI426222305
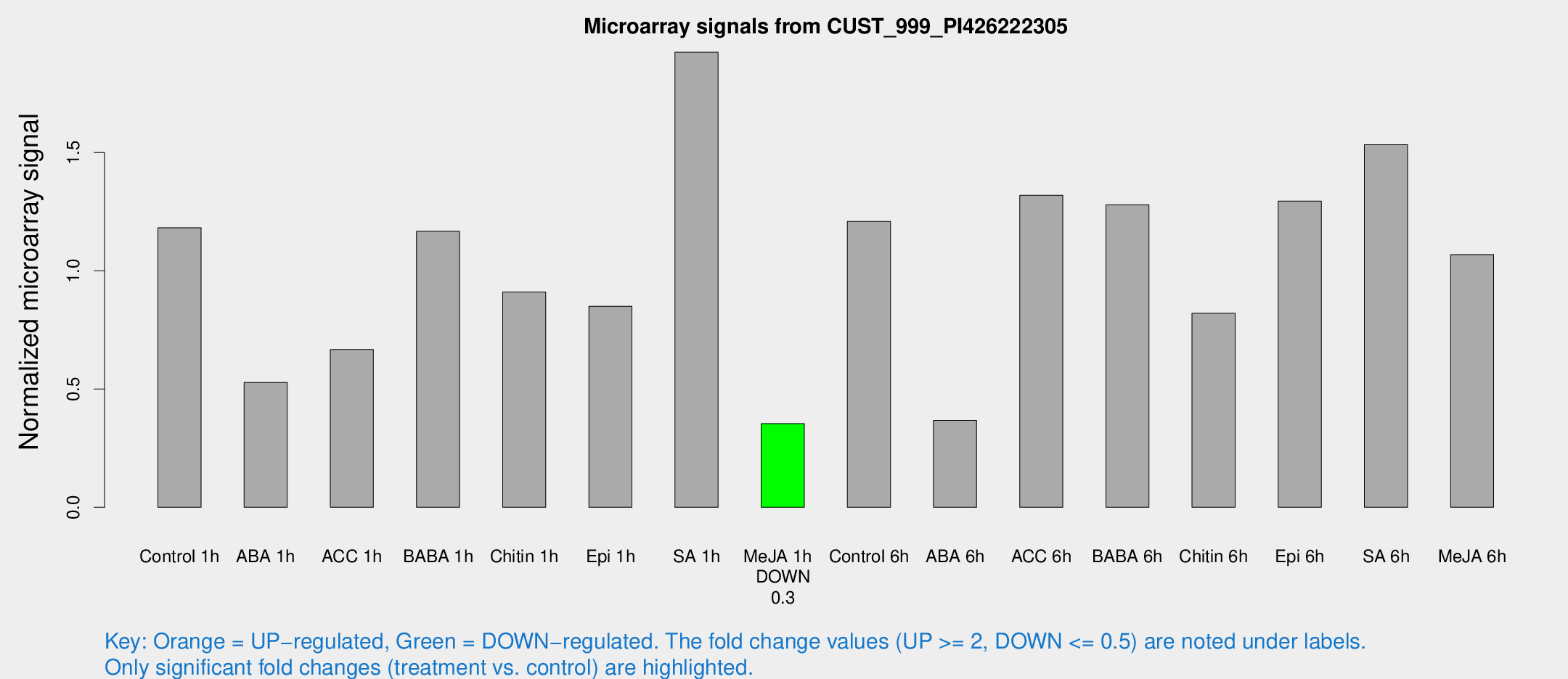
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 100.621 | 18.1325 | 1.18207 | 0.131153 |
| ABA 1h | 41.8835 | 10.8324 | 0.527794 | 0.212192 |
| ACC 1h | 70.723 | 25.6455 | 0.667101 | 0.35662 |
| BABA 1h | 96.2779 | 17.0017 | 1.16776 | 0.108218 |
| Chitin 1h | 68.581 | 8.29507 | 0.910764 | 0.071621 |
| Epi 1h | 60.9531 | 5.01755 | 0.850193 | 0.0696965 |
| SA 1h | 170.933 | 33.7554 | 1.92428 | 0.338781 |
| Me-JA 1h | 24.6414 | 4.71481 | 0.353819 | 0.0605354 |
| Control 6h | 110.813 | 31.4998 | 1.20914 | 0.320555 |
| ABA 6h | 43.1703 | 19.3929 | 0.367657 | 0.251233 |
| ACC 6h | 125.071 | 8.5222 | 1.31906 | 0.198356 |
| BABA 6h | 121.827 | 19.9068 | 1.2794 | 0.1887 |
| Chitin 6h | 76.8883 | 20.501 | 0.821103 | 0.237234 |
| Epi 6h | 125.187 | 22.7729 | 1.29464 | 0.188391 |
| SA 6h | 125.536 | 8.36563 | 1.5332 | 0.184875 |
| Me-JA 6h | 104.418 | 45.3713 | 1.06852 | 0.383859 |
Source Transcript PGSC0003DMT400003463 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | Solyc02g089090.2 | +1 | 4e-49 | 171 | 92/134 (69%) | genomic_reference:SL2.50ch02 gene_region:45568082-45571263 transcript_region:SL2.50ch02:45568082..45571263+ go_terms:GO:0004672 functional_description:Pto-like, Serine/threonine kinase protein, resistance protein |
| TAIR PP10 | AT5G39030.1 | +1 | 4e-26 | 106 | 69/150 (46%) | Protein kinase superfamily protein | chr5:15620066-15622486 FORWARD LENGTH=806 |
