Probe CUST_9416_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_9416_PI426222305 | JHI_St_60k_v1 | DMT400006434 | GCATGATCTGATATGCTCAGTGTACTCTCTTCTATTATCATATTGTATTGTTGCTGTTGA |
All Microarray Probes Designed to Gene DMG400002512
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_9332_PI426222305 | JHI_St_60k_v1 | DMT400006435 | TTTCAAGGCAGGCAAGTTCCTAGTCAGGTAAAGTGAACAGATTCCTTTCCTTTCTAATTG |
| CUST_9416_PI426222305 | JHI_St_60k_v1 | DMT400006434 | GCATGATCTGATATGCTCAGTGTACTCTCTTCTATTATCATATTGTATTGTTGCTGTTGA |
| CUST_9390_PI426222305 | JHI_St_60k_v1 | DMT400006436 | GCATGATCTGATATGCTCAGTGTACTCTCTTCTATTATCATATTGTATTGTTGCTGTTGA |
Microarray Signals from CUST_9416_PI426222305
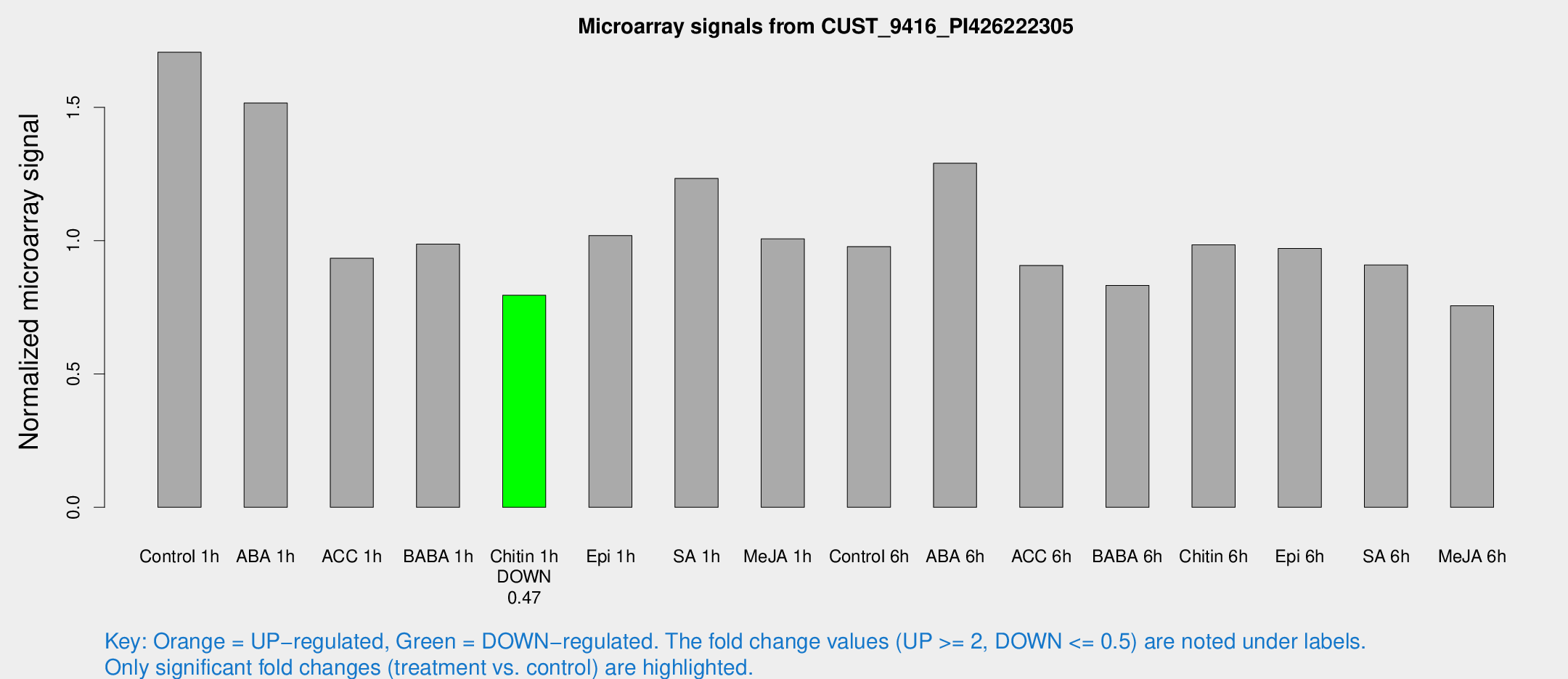
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 370.028 | 26.9236 | 1.70672 | 0.0996511 |
| ABA 1h | 292.752 | 30.861 | 1.51604 | 0.11934 |
| ACC 1h | 241.26 | 79.6153 | 0.93408 | 0.358204 |
| BABA 1h | 211.742 | 36.6126 | 0.987024 | 0.0920428 |
| Chitin 1h | 155.076 | 11.2397 | 0.795074 | 0.0903941 |
| Epi 1h | 192.783 | 21.7263 | 1.01914 | 0.114271 |
| SA 1h | 278.633 | 39.8999 | 1.23343 | 0.173173 |
| Me-JA 1h | 179.306 | 19.8275 | 1.00651 | 0.0609337 |
| Control 6h | 217.621 | 34.4905 | 0.977854 | 0.0789097 |
| ABA 6h | 299.973 | 35.3488 | 1.29014 | 0.0793248 |
| ACC 6h | 228.122 | 29.8632 | 0.907201 | 0.0546847 |
| BABA 6h | 202.345 | 16.9646 | 0.832381 | 0.0672534 |
| Chitin 6h | 226.025 | 13.5868 | 0.984287 | 0.0591672 |
| Epi 6h | 236.256 | 14.2004 | 0.970473 | 0.0731988 |
| SA 6h | 202.002 | 37.5319 | 0.908882 | 0.0842233 |
| Me-JA 6h | 165.633 | 24.0168 | 0.75611 | 0.0600829 |
Source Transcript PGSC0003DMT400006434 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G11730.1 | +2 | 5e-167 | 488 | 236/379 (62%) | Core-2/I-branching beta-1,6-N-acetylglucosaminyltransferase family protein | chr5:3780963-3782473 FORWARD LENGTH=386 |
