Probe CUST_9390_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_9390_PI426222305 | JHI_St_60k_v1 | DMT400006436 | GCATGATCTGATATGCTCAGTGTACTCTCTTCTATTATCATATTGTATTGTTGCTGTTGA |
All Microarray Probes Designed to Gene DMG400002512
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_9332_PI426222305 | JHI_St_60k_v1 | DMT400006435 | TTTCAAGGCAGGCAAGTTCCTAGTCAGGTAAAGTGAACAGATTCCTTTCCTTTCTAATTG |
| CUST_9416_PI426222305 | JHI_St_60k_v1 | DMT400006434 | GCATGATCTGATATGCTCAGTGTACTCTCTTCTATTATCATATTGTATTGTTGCTGTTGA |
| CUST_9390_PI426222305 | JHI_St_60k_v1 | DMT400006436 | GCATGATCTGATATGCTCAGTGTACTCTCTTCTATTATCATATTGTATTGTTGCTGTTGA |
Microarray Signals from CUST_9390_PI426222305
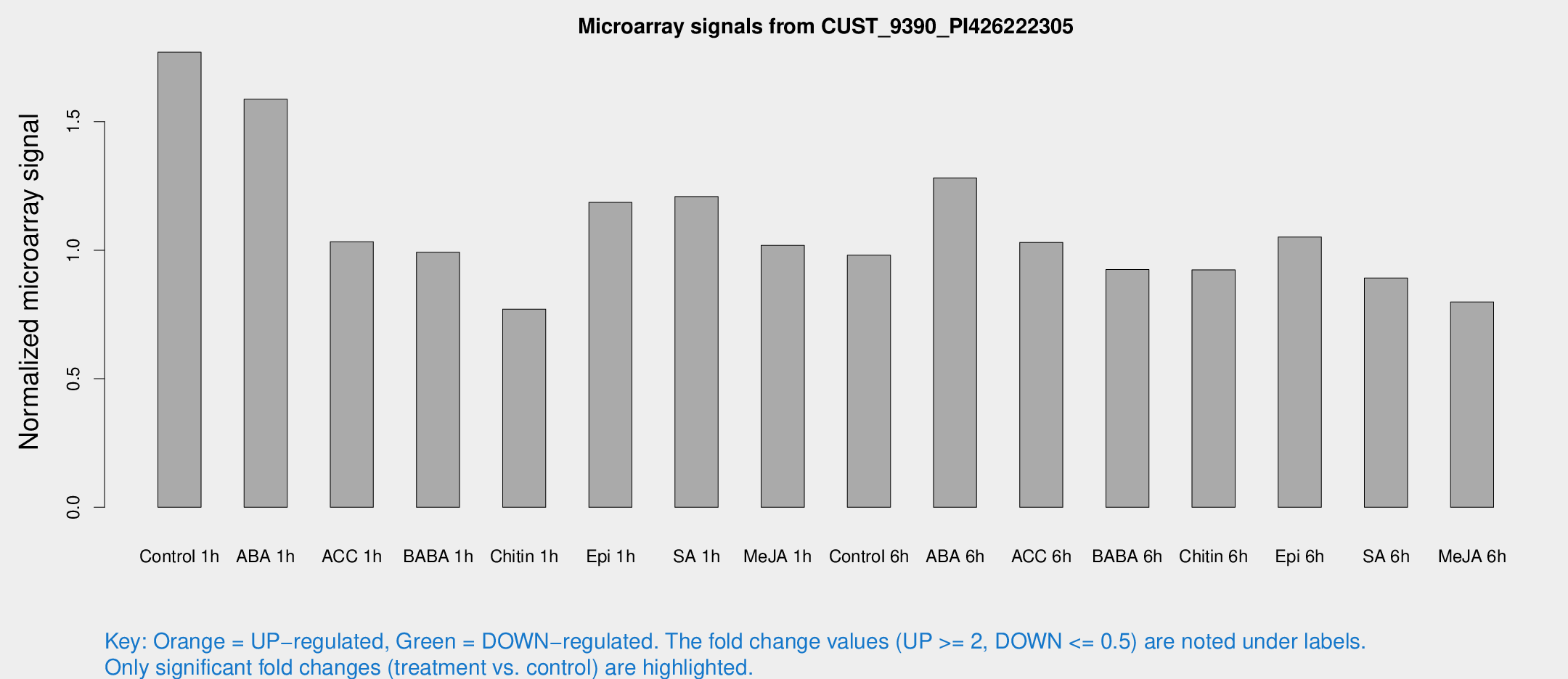
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 361.657 | 21.1618 | 1.76997 | 0.130901 |
| ABA 1h | 290.822 | 36.5135 | 1.58716 | 0.174731 |
| ACC 1h | 252.742 | 85.9362 | 1.03303 | 0.398897 |
| BABA 1h | 203.691 | 44.2511 | 0.992034 | 0.151827 |
| Chitin 1h | 141.415 | 8.74362 | 0.770678 | 0.0475788 |
| Epi 1h | 213.552 | 31.7753 | 1.18599 | 0.183253 |
| SA 1h | 256.533 | 30.8765 | 1.2087 | 0.165715 |
| Me-JA 1h | 170.399 | 12.2901 | 1.01854 | 0.0618937 |
| Control 6h | 202.895 | 22.0617 | 0.98058 | 0.0588404 |
| ABA 6h | 280.406 | 28.2922 | 1.28102 | 0.0755407 |
| ACC 6h | 243.03 | 22.8992 | 1.03019 | 0.0927842 |
| BABA 6h | 211.946 | 13.9014 | 0.924833 | 0.0556232 |
| Chitin 6h | 200.915 | 12.1593 | 0.92325 | 0.0654917 |
| Epi 6h | 243.903 | 24.5996 | 1.0512 | 0.119156 |
| SA 6h | 187.68 | 36.054 | 0.891531 | 0.0903294 |
| Me-JA 6h | 164.52 | 20.6144 | 0.798772 | 0.0634516 |
Source Transcript PGSC0003DMT400006436 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G11730.1 | +2 | 2e-115 | 344 | 154/211 (73%) | Core-2/I-branching beta-1,6-N-acetylglucosaminyltransferase family protein | chr5:3780963-3782473 FORWARD LENGTH=386 |
