Probe CUST_8367_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_8367_PI426222305 | JHI_St_60k_v1 | DMT400075463 | GAAGCAGTAGATGAAGGAGGGAGTTCTGATAAGAATATTGAAGAATTTCTTTCAAACTTG |
All Microarray Probes Designed to Gene DMG400029350
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_8381_PI426222305 | JHI_St_60k_v1 | DMT400075467 | GAGTGCTGCATAAATTAGTTCTATGTGGTCTGTTTATGAAAAAGACCTACATTACAGTGA |
| CUST_8367_PI426222305 | JHI_St_60k_v1 | DMT400075463 | GAAGCAGTAGATGAAGGAGGGAGTTCTGATAAGAATATTGAAGAATTTCTTTCAAACTTG |
| CUST_8244_PI426222305 | JHI_St_60k_v1 | DMT400075464 | GAAGCAGTAGATGAAGGAGGGAGTTCTGATAAGAATATTGAAGAATTTCTTTCAAACTTG |
| CUST_8049_PI426222305 | JHI_St_60k_v1 | DMT400075462 | CTAACAGTGAAACCTTTGGATGTTTCTTTACTCACTGTGGATGGAATTCGACACTGGAAG |
Microarray Signals from CUST_8367_PI426222305
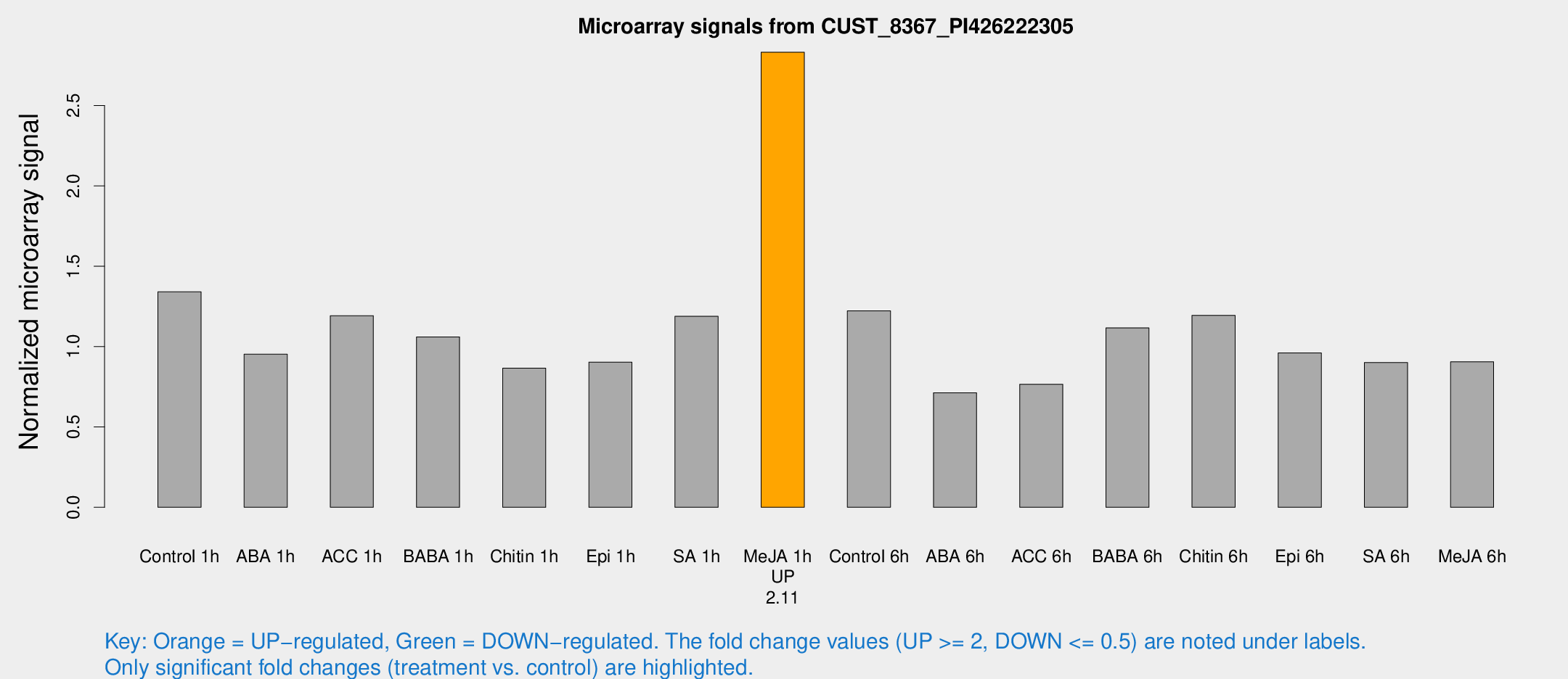
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 732.822 | 111.348 | 1.34052 | 0.117743 |
| ABA 1h | 465.152 | 84.5109 | 0.952588 | 0.126056 |
| ACC 1h | 688.772 | 142.969 | 1.19181 | 0.194223 |
| BABA 1h | 547.159 | 31.8078 | 1.05989 | 0.086712 |
| Chitin 1h | 422.182 | 49.7114 | 0.865612 | 0.0980857 |
| Epi 1h | 418.614 | 24.3918 | 0.903192 | 0.0525896 |
| SA 1h | 675.179 | 126.451 | 1.18809 | 0.206926 |
| Me-JA 1h | 1238.3 | 71.6641 | 2.83116 | 0.163634 |
| Control 6h | 716.831 | 185.246 | 1.22261 | 0.283754 |
| ABA 6h | 413.427 | 59.8992 | 0.712634 | 0.0693094 |
| ACC 6h | 480.265 | 68.9846 | 0.765632 | 0.0555812 |
| BABA 6h | 683.959 | 107.312 | 1.11593 | 0.157986 |
| Chitin 6h | 680.959 | 39.5263 | 1.19441 | 0.0692661 |
| Epi 6h | 592.837 | 86.311 | 0.960407 | 0.207145 |
| SA 6h | 508.806 | 121.393 | 0.900258 | 0.140535 |
| Me-JA 6h | 514.163 | 121.061 | 0.905488 | 0.162801 |
Source Transcript PGSC0003DMT400075463 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G05680.1 | +2 | 5e-153 | 448 | 227/447 (51%) | Uridine diphosphate glycosyltransferase 74E2 | chr1:1703196-1704639 REVERSE LENGTH=453 |
