Probe CUST_8049_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_8049_PI426222305 | JHI_St_60k_v1 | DMT400075462 | CTAACAGTGAAACCTTTGGATGTTTCTTTACTCACTGTGGATGGAATTCGACACTGGAAG |
All Microarray Probes Designed to Gene DMG400029350
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_8381_PI426222305 | JHI_St_60k_v1 | DMT400075467 | GAGTGCTGCATAAATTAGTTCTATGTGGTCTGTTTATGAAAAAGACCTACATTACAGTGA |
| CUST_8367_PI426222305 | JHI_St_60k_v1 | DMT400075463 | GAAGCAGTAGATGAAGGAGGGAGTTCTGATAAGAATATTGAAGAATTTCTTTCAAACTTG |
| CUST_8244_PI426222305 | JHI_St_60k_v1 | DMT400075464 | GAAGCAGTAGATGAAGGAGGGAGTTCTGATAAGAATATTGAAGAATTTCTTTCAAACTTG |
| CUST_8049_PI426222305 | JHI_St_60k_v1 | DMT400075462 | CTAACAGTGAAACCTTTGGATGTTTCTTTACTCACTGTGGATGGAATTCGACACTGGAAG |
Microarray Signals from CUST_8049_PI426222305
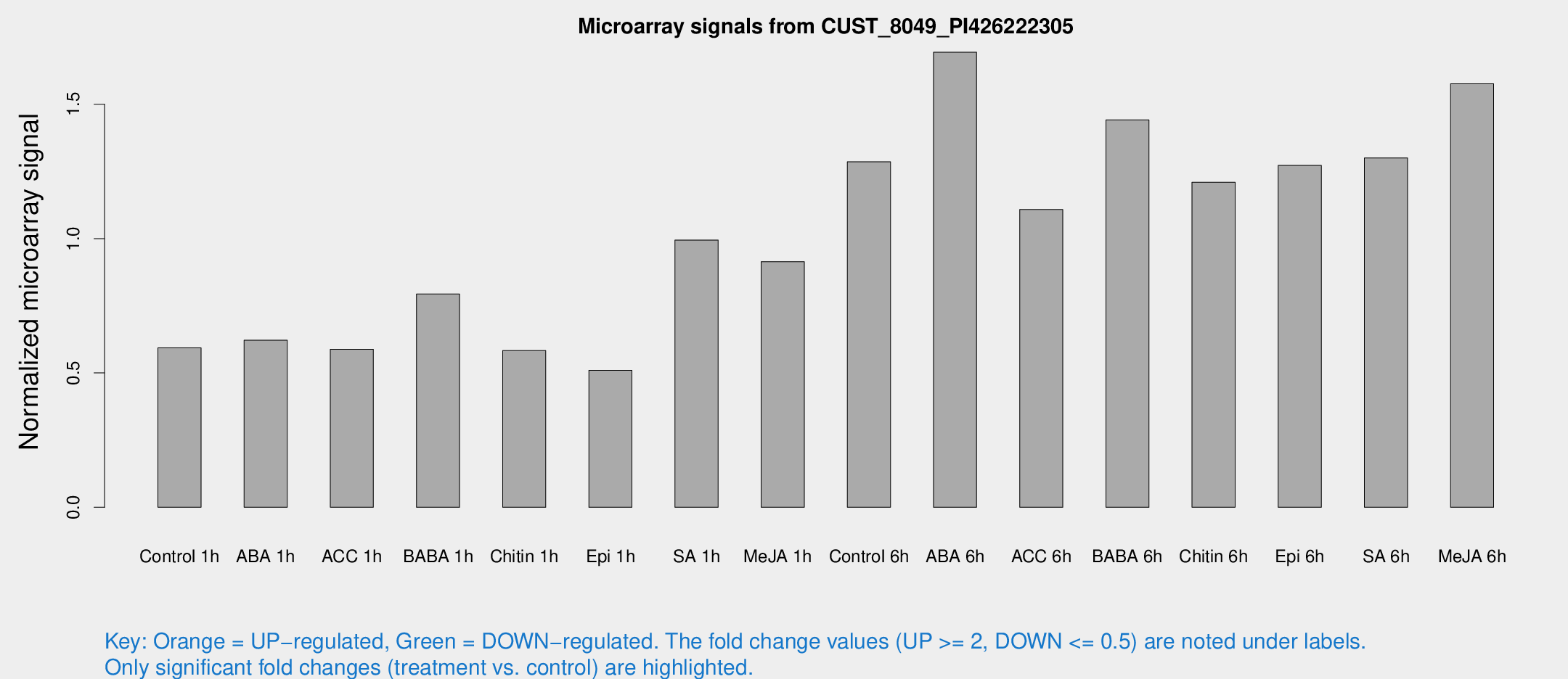
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 64.7932 | 12.0854 | 0.593312 | 0.12073 |
| ABA 1h | 59.5444 | 9.76475 | 0.621862 | 0.0750903 |
| ACC 1h | 70.5993 | 20.6522 | 0.587919 | 0.170667 |
| BABA 1h | 84.7684 | 17.7081 | 0.793831 | 0.109985 |
| Chitin 1h | 55.3347 | 4.37028 | 0.5832 | 0.0468656 |
| Epi 1h | 48.5542 | 9.61141 | 0.509804 | 0.109848 |
| SA 1h | 111.016 | 21.3156 | 0.99458 | 0.145251 |
| Me-JA 1h | 84.1659 | 23.611 | 0.913985 | 0.187523 |
| Control 6h | 142.822 | 30.1393 | 1.28565 | 0.19252 |
| ABA 6h | 199.499 | 45.1816 | 1.69347 | 0.312637 |
| ACC 6h | 134.976 | 14.136 | 1.10824 | 0.07072 |
| BABA 6h | 170.819 | 15.1742 | 1.44142 | 0.0882822 |
| Chitin 6h | 138.135 | 21.0246 | 1.20961 | 0.140283 |
| Epi 6h | 155.105 | 25.0683 | 1.2725 | 0.157013 |
| SA 6h | 145.689 | 35.4355 | 1.30023 | 0.22881 |
| Me-JA 6h | 169.492 | 28.2318 | 1.57595 | 0.133703 |
Source Transcript PGSC0003DMT400075462 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G05680.1 | +1 | 2e-45 | 155 | 72/110 (65%) | Uridine diphosphate glycosyltransferase 74E2 | chr1:1703196-1704639 REVERSE LENGTH=453 |
