Probe CUST_7803_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_7803_PI426222305 | JHI_St_60k_v1 | DMT400025571 | TATGGTACAGATATGTCTGTCTTTCTCCTGAATATCAACAAGATCTCCGTTTCTTATCTT |
All Microarray Probes Designed to Gene DMG400009880
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_7673_PI426222305 | JHI_St_60k_v1 | DMT400025573 | AGAGGAGCTGGTTTGTAGTTAGCTATGGTAACAGTTGGCAGAACTGATGCAACTGGTTTC |
| CUST_7762_PI426222305 | JHI_St_60k_v1 | DMT400025572 | GATTCTTGTCCAACATTTAAGTAGCTTCTTCTGATATATTCCACATGCAAAAGTCCAACA |
| CUST_7803_PI426222305 | JHI_St_60k_v1 | DMT400025571 | TATGGTACAGATATGTCTGTCTTTCTCCTGAATATCAACAAGATCTCCGTTTCTTATCTT |
| CUST_7595_PI426222305 | JHI_St_60k_v1 | DMT400025574 | AGAGGAGCTGGTTTGTAGTTAGCTATGGTAACAGTTGGCAGAACTGATGCAACTGGTTTC |
Microarray Signals from CUST_7803_PI426222305
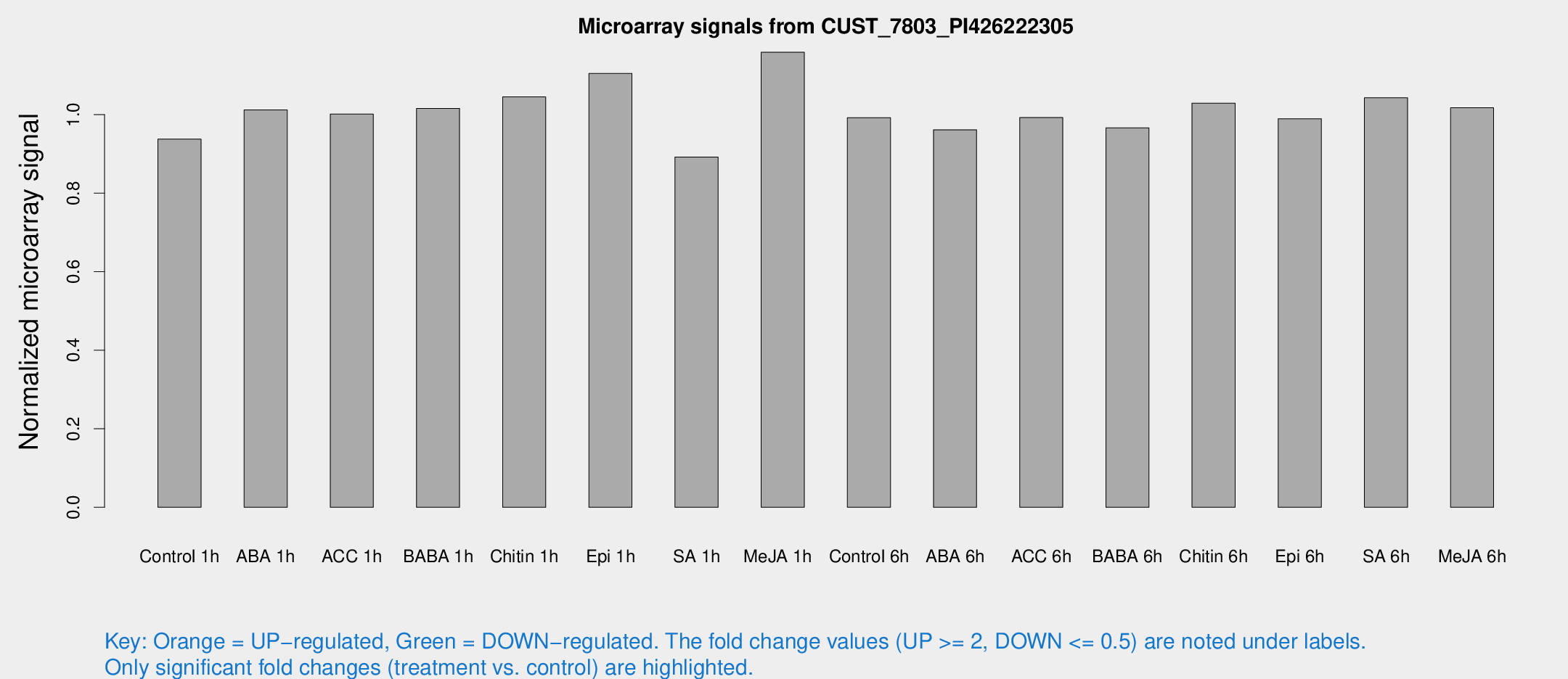
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 5.44509 | 3.156 | 0.937776 | 0.543022 |
| ABA 1h | 5.19416 | 3.00872 | 1.01197 | 0.586036 |
| ACC 1h | 5.97389 | 3.46676 | 1.00111 | 0.580002 |
| BABA 1h | 5.68697 | 3.29761 | 1.01574 | 0.588184 |
| Chitin 1h | 5.41757 | 3.15475 | 1.04526 | 0.599135 |
| Epi 1h | 5.54713 | 3.10612 | 1.10462 | 0.61734 |
| SA 1h | 5.31995 | 3.08621 | 0.891804 | 0.516824 |
| Me-JA 1h | 5.51122 | 3.20579 | 1.15881 | 0.671116 |
| Control 6h | 5.77268 | 3.34587 | 0.992273 | 0.574767 |
| ABA 6h | 5.93226 | 3.43706 | 0.961352 | 0.55668 |
| ACC 6h | 6.74743 | 3.99654 | 0.992424 | 0.574683 |
| BABA 6h | 6.29195 | 3.65479 | 0.966276 | 0.559698 |
| Chitin 6h | 6.35714 | 3.68538 | 1.02897 | 0.59617 |
| Epi 6h | 6.53632 | 3.82346 | 0.989431 | 0.572956 |
| SA 6h | 5.9836 | 3.46752 | 1.04269 | 0.604195 |
| Me-JA 6h | 5.88903 | 3.30524 | 1.01729 | 0.571271 |
Source Transcript PGSC0003DMT400025571 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G75890.1 | +1 | 5e-82 | 268 | 138/281 (49%) | GDSL-like Lipase/Acylhydrolase superfamily protein | chr1:28493051-28495047 FORWARD LENGTH=379 |
