Probe CUST_7762_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_7762_PI426222305 | JHI_St_60k_v1 | DMT400025572 | GATTCTTGTCCAACATTTAAGTAGCTTCTTCTGATATATTCCACATGCAAAAGTCCAACA |
All Microarray Probes Designed to Gene DMG400009880
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_7673_PI426222305 | JHI_St_60k_v1 | DMT400025573 | AGAGGAGCTGGTTTGTAGTTAGCTATGGTAACAGTTGGCAGAACTGATGCAACTGGTTTC |
| CUST_7762_PI426222305 | JHI_St_60k_v1 | DMT400025572 | GATTCTTGTCCAACATTTAAGTAGCTTCTTCTGATATATTCCACATGCAAAAGTCCAACA |
| CUST_7803_PI426222305 | JHI_St_60k_v1 | DMT400025571 | TATGGTACAGATATGTCTGTCTTTCTCCTGAATATCAACAAGATCTCCGTTTCTTATCTT |
| CUST_7595_PI426222305 | JHI_St_60k_v1 | DMT400025574 | AGAGGAGCTGGTTTGTAGTTAGCTATGGTAACAGTTGGCAGAACTGATGCAACTGGTTTC |
Microarray Signals from CUST_7762_PI426222305
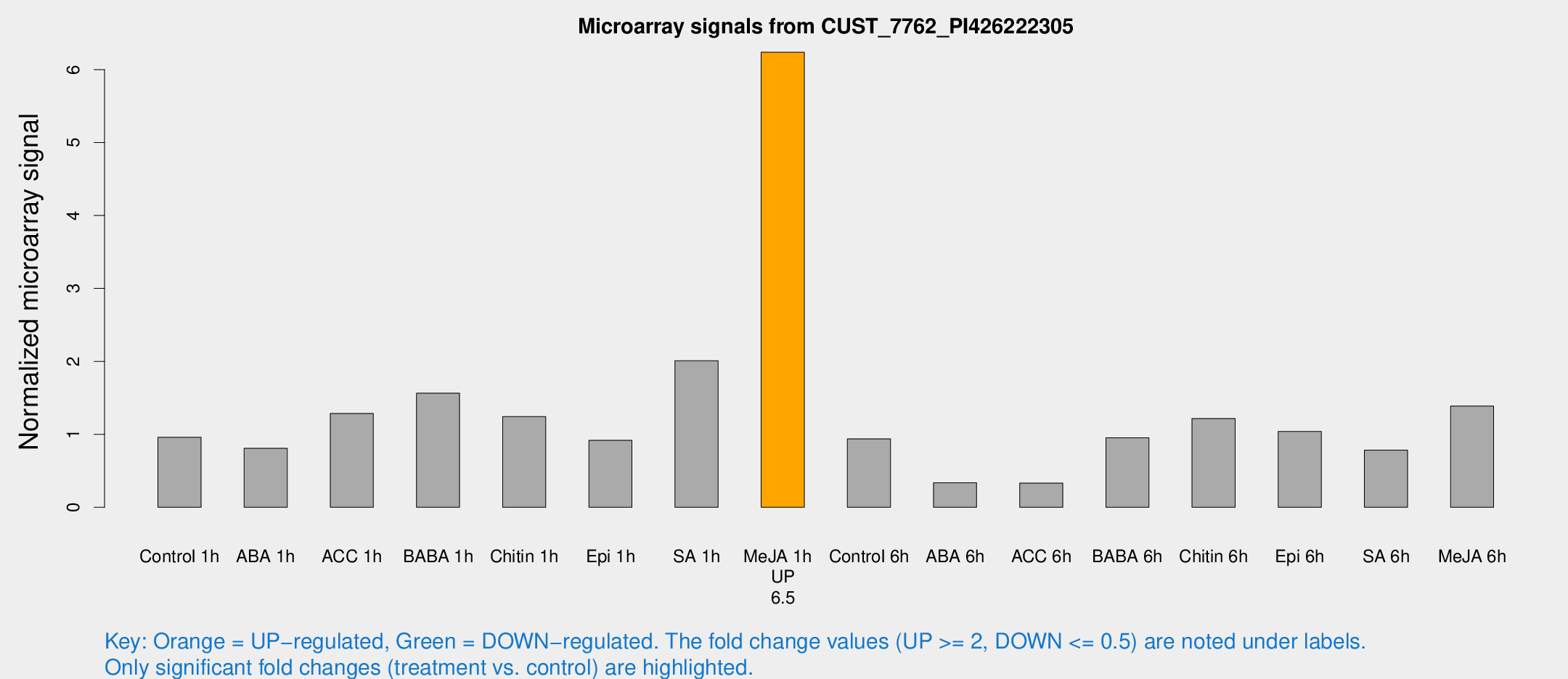
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 107.481 | 13.1308 | 0.95957 | 0.0682899 |
| ABA 1h | 81.1169 | 12.8173 | 0.809436 | 0.0734396 |
| ACC 1h | 159.033 | 46.4502 | 1.28655 | 0.320503 |
| BABA 1h | 166.903 | 10.3669 | 1.56494 | 0.148902 |
| Chitin 1h | 127.39 | 22.006 | 1.24463 | 0.289088 |
| Epi 1h | 90.2865 | 14.8698 | 0.91868 | 0.154 |
| SA 1h | 276.728 | 119.856 | 2.0083 | 1.12822 |
| Me-JA 1h | 564.386 | 33.3761 | 6.23784 | 0.362369 |
| Control 6h | 121.47 | 43.4107 | 0.938272 | 0.327941 |
| ABA 6h | 45.5371 | 17.0342 | 0.337181 | 0.155862 |
| ACC 6h | 44.4882 | 9.32327 | 0.332637 | 0.077724 |
| BABA 6h | 119.335 | 13.1873 | 0.952659 | 0.0724945 |
| Chitin 6h | 150.129 | 34.4952 | 1.21657 | 0.269373 |
| Epi 6h | 137.076 | 29.9887 | 1.03872 | 0.346591 |
| SA 6h | 85.8326 | 6.36341 | 0.783321 | 0.135309 |
| Me-JA 6h | 158.466 | 28.3356 | 1.38826 | 0.166247 |
Source Transcript PGSC0003DMT400025572 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G75900.1 | +2 | 5e-90 | 280 | 150/287 (52%) | GDSL-like Lipase/Acylhydrolase superfamily protein | chr1:28499179-28500943 FORWARD LENGTH=364 |
