Probe CUST_7416_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_7416_PI426222305 | JHI_St_60k_v1 | DMT400025713 | GGGATGTAAATAATAAGGAAGCAAGTATGTTACTTGTTATTTATTGTGTGACACGCATCC |
All Microarray Probes Designed to Gene DMG401009929
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_7416_PI426222305 | JHI_St_60k_v1 | DMT400025713 | GGGATGTAAATAATAAGGAAGCAAGTATGTTACTTGTTATTTATTGTGTGACACGCATCC |
| CUST_7841_PI426222305 | JHI_St_60k_v1 | DMT400025715 | GGGATGTAAATAATAAGGAAGCAAGTATGTTACTTGTTATTTATTGTGTGACACGCATCC |
| CUST_7410_PI426222305 | JHI_St_60k_v1 | DMT400025716 | GTTGTTGTTGCTTCCTTGTTCTCAGGTTTCCGAGTAAACATAATTTGGATTGATTTTACA |
| CUST_7383_PI426222305 | JHI_St_60k_v1 | DMT400025714 | GGGATGTAAATAATAAGGAAGCAAGTATGTTACTTGTTATTTATTGTGTGACACGCATCC |
Microarray Signals from CUST_7416_PI426222305
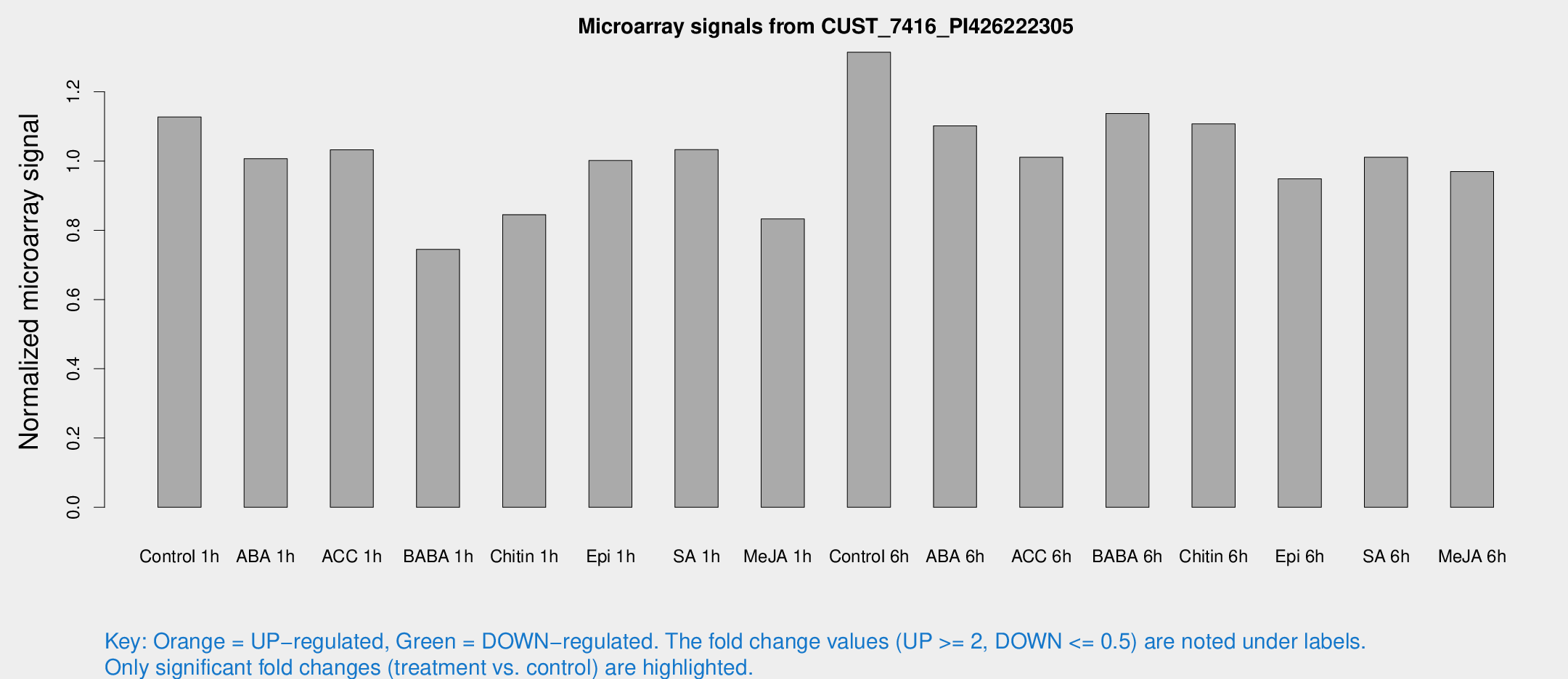
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 735.742 | 94.5067 | 1.12699 | 0.0759322 |
| ABA 1h | 572.752 | 33.2406 | 1.00686 | 0.0584131 |
| ACC 1h | 689.564 | 73.9851 | 1.03248 | 0.0660977 |
| BABA 1h | 492.145 | 111.538 | 0.744633 | 0.122854 |
| Chitin 1h | 491.081 | 35.5754 | 0.844898 | 0.0491274 |
| Epi 1h | 567.14 | 76.185 | 1.00132 | 0.109377 |
| SA 1h | 690.37 | 76.46 | 1.03274 | 0.0598507 |
| Me-JA 1h | 440.728 | 41.1726 | 0.832778 | 0.0485259 |
| Control 6h | 869.432 | 137.571 | 1.31403 | 0.0984556 |
| ABA 6h | 788.263 | 171.197 | 1.10163 | 0.176376 |
| ACC 6h | 765.205 | 118.257 | 1.01075 | 0.099845 |
| BABA 6h | 822.319 | 59.3514 | 1.13708 | 0.084638 |
| Chitin 6h | 772.796 | 109.905 | 1.1071 | 0.121364 |
| Epi 6h | 688.543 | 40.014 | 0.948744 | 0.107182 |
| SA 6h | 717.706 | 206.649 | 1.0108 | 0.251231 |
| Me-JA 6h | 640.644 | 114.215 | 0.969635 | 0.0922513 |
Source Transcript PGSC0003DMT400025713 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G76570.1 | +2 | 1e-165 | 479 | 224/280 (80%) | Chlorophyll A-B binding family protein | chr1:28729132-28730754 FORWARD LENGTH=327 |
