Probe CUST_7410_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_7410_PI426222305 | JHI_St_60k_v1 | DMT400025716 | GTTGTTGTTGCTTCCTTGTTCTCAGGTTTCCGAGTAAACATAATTTGGATTGATTTTACA |
All Microarray Probes Designed to Gene DMG401009929
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_7416_PI426222305 | JHI_St_60k_v1 | DMT400025713 | GGGATGTAAATAATAAGGAAGCAAGTATGTTACTTGTTATTTATTGTGTGACACGCATCC |
| CUST_7841_PI426222305 | JHI_St_60k_v1 | DMT400025715 | GGGATGTAAATAATAAGGAAGCAAGTATGTTACTTGTTATTTATTGTGTGACACGCATCC |
| CUST_7410_PI426222305 | JHI_St_60k_v1 | DMT400025716 | GTTGTTGTTGCTTCCTTGTTCTCAGGTTTCCGAGTAAACATAATTTGGATTGATTTTACA |
| CUST_7383_PI426222305 | JHI_St_60k_v1 | DMT400025714 | GGGATGTAAATAATAAGGAAGCAAGTATGTTACTTGTTATTTATTGTGTGACACGCATCC |
Microarray Signals from CUST_7410_PI426222305
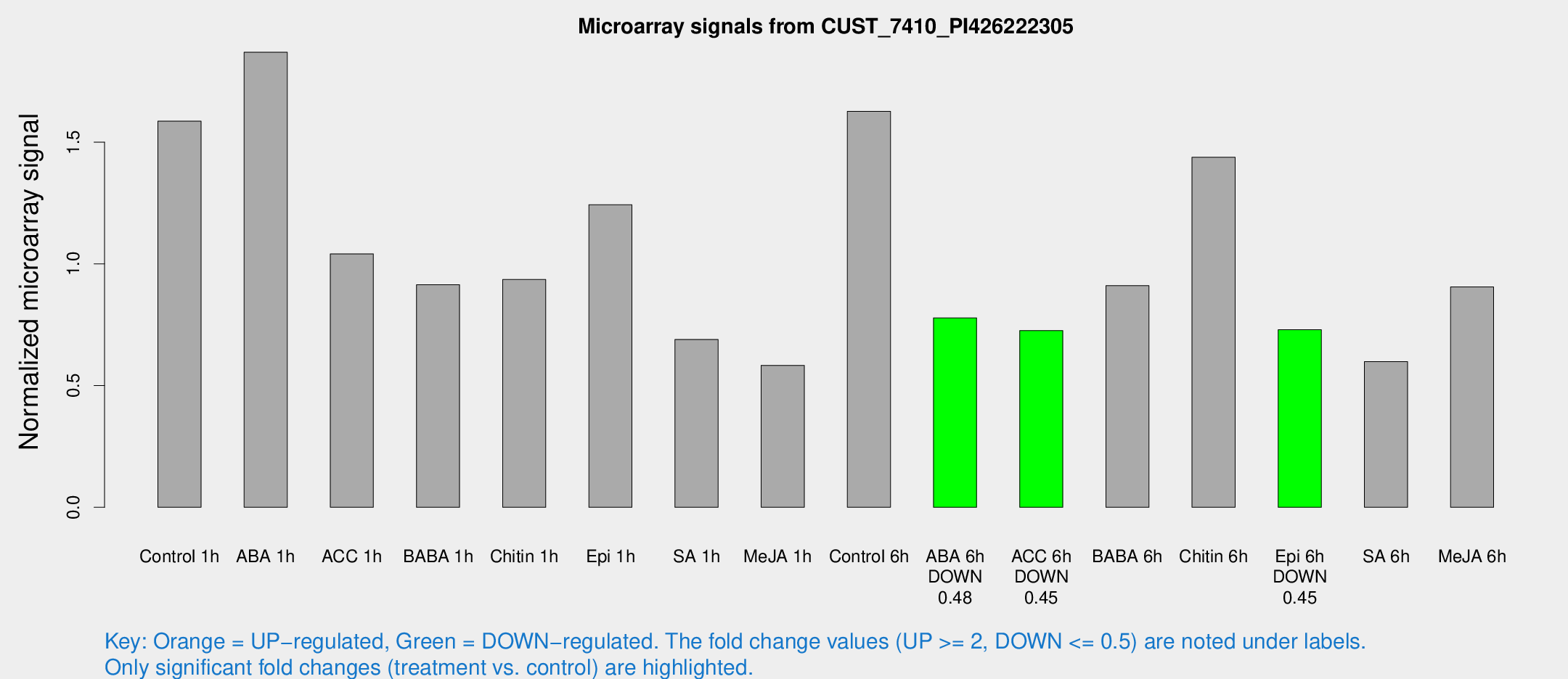
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 63.3713 | 4.8625 | 1.58638 | 0.125228 |
| ABA 1h | 71.4552 | 18.6655 | 1.86957 | 0.700089 |
| ACC 1h | 48.1662 | 14.5783 | 1.04094 | 0.329493 |
| BABA 1h | 35.9612 | 5.0699 | 0.914518 | 0.186814 |
| Chitin 1h | 34.2947 | 4.88258 | 0.935986 | 0.181435 |
| Epi 1h | 43.2838 | 4.0608 | 1.2429 | 0.118449 |
| SA 1h | 29.9908 | 7.3036 | 0.689672 | 0.149878 |
| Me-JA 1h | 21.0834 | 5.82813 | 0.58323 | 0.234665 |
| Control 6h | 65.333 | 5.14475 | 1.62674 | 0.128692 |
| ABA 6h | 34.356 | 6.26963 | 0.778342 | 0.13276 |
| ACC 6h | 33.7891 | 4.5617 | 0.726068 | 0.0972717 |
| BABA 6h | 41.4863 | 5.70553 | 0.910507 | 0.10652 |
| Chitin 6h | 62.3035 | 8.1012 | 1.43814 | 0.202535 |
| Epi 6h | 33.4452 | 4.57589 | 0.729402 | 0.0988966 |
| SA 6h | 24.9419 | 5.58418 | 0.598495 | 0.222163 |
| Me-JA 6h | 36.7859 | 5.20381 | 0.905234 | 0.104037 |
Source Transcript PGSC0003DMT400025716 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G76570.1 | +1 | 5e-18 | 85 | 34/51 (67%) | Chlorophyll A-B binding family protein | chr1:28729132-28730754 FORWARD LENGTH=327 |
