Probe CUST_5785_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_5785_PI426222305 | JHI_St_60k_v1 | DMT400023102 | CTTTATGATGTGGTAAAGGGATATCATTGTTAAATACTCTGAGGAAGTGAAGAAACTCGG |
All Microarray Probes Designed to Gene DMG400008947
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_5694_PI426222305 | JHI_St_60k_v1 | DMT400023103 | CCTGACACAAACATCAGTGCGTCAAAACAACAGTACAATGATTAATTTGAGTTATTCTCT |
| CUST_5507_PI426222305 | JHI_St_60k_v1 | DMT400023104 | GACTTGATGGGACTTCTGCTCTACAACATTTTCGACTTTAAACTATCTTTTCATTTGTCT |
| CUST_5785_PI426222305 | JHI_St_60k_v1 | DMT400023102 | CTTTATGATGTGGTAAAGGGATATCATTGTTAAATACTCTGAGGAAGTGAAGAAACTCGG |
Microarray Signals from CUST_5785_PI426222305
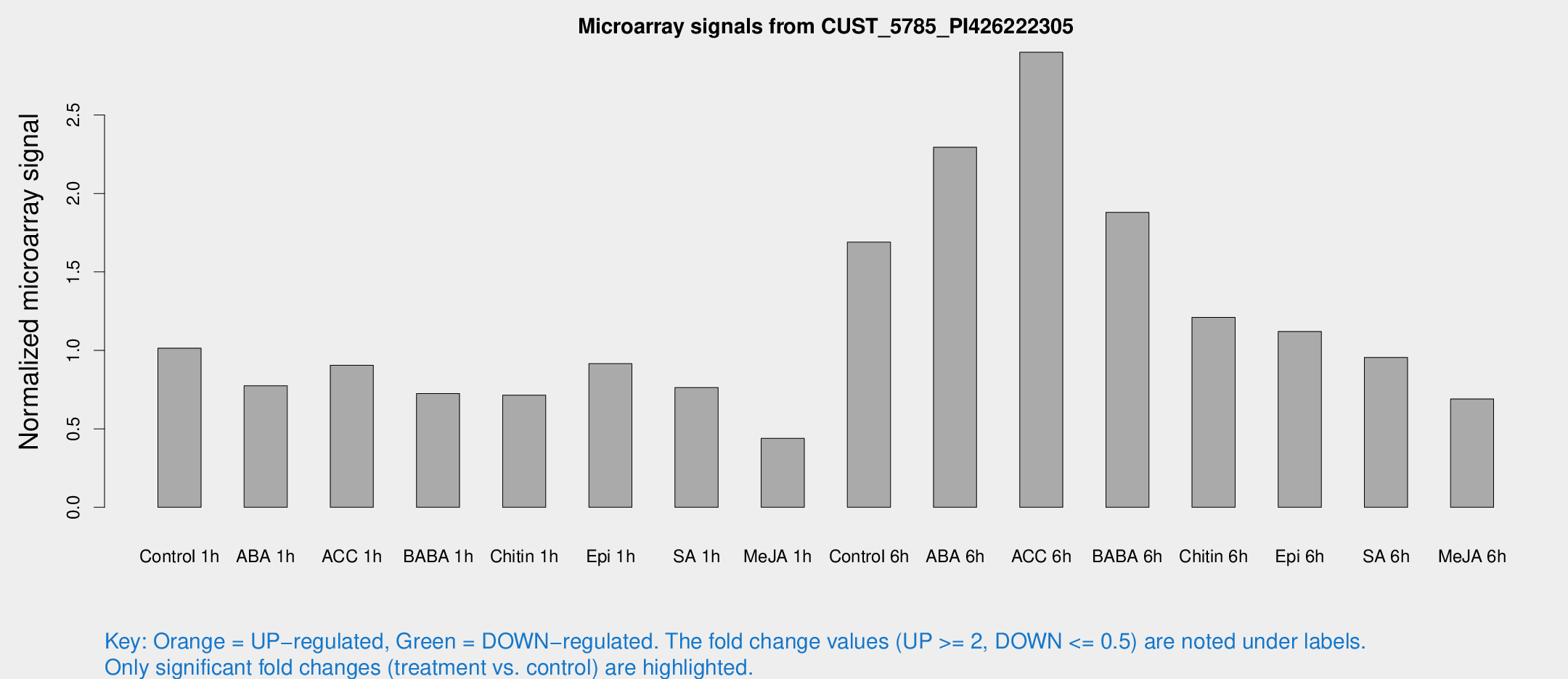
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 54.7675 | 13.3546 | 1.01409 | 0.176138 |
| ABA 1h | 36.5232 | 6.86368 | 0.774219 | 0.131179 |
| ACC 1h | 49.6273 | 10.4478 | 0.904898 | 0.147611 |
| BABA 1h | 39.609 | 11.1085 | 0.725076 | 0.177968 |
| Chitin 1h | 37.6515 | 14.4156 | 0.715096 | 0.21464 |
| Epi 1h | 41.9569 | 7.14868 | 0.915655 | 0.165149 |
| SA 1h | 54.8384 | 31.3888 | 0.763486 | 0.448484 |
| Me-JA 1h | 18.8342 | 3.36233 | 0.440064 | 0.0827769 |
| Control 6h | 98.7993 | 31.5525 | 1.68957 | 0.541417 |
| ABA 6h | 140.848 | 41.0983 | 2.29467 | 0.820783 |
| ACC 6h | 171.73 | 15.6036 | 2.90004 | 0.246268 |
| BABA 6h | 111.42 | 20.4025 | 1.87966 | 0.279562 |
| Chitin 6h | 71.329 | 20.953 | 1.21081 | 0.306403 |
| Epi 6h | 65.828 | 8.78911 | 1.12066 | 0.123137 |
| SA 6h | 57.8828 | 19.5781 | 0.955458 | 0.366231 |
| Me-JA 6h | 40.008 | 11.9526 | 0.690523 | 0.230728 |
Source Transcript PGSC0003DMT400023102 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G06620.1 | +2 | 8e-86 | 276 | 126/190 (66%) | 2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein | chr1:2025618-2027094 FORWARD LENGTH=365 |
