Probe CUST_52483_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_52483_PI426222305 | JHI_St_60k_v1 | DMT400049092 | TGAGTTTTTTCGTCAAACTCAGTCCGAGGAGATCTTTACAGGAATTGTGAGAGAAGTGCA |
All Microarray Probes Designed to Gene DMG400019082
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_52484_PI426222305 | JHI_St_60k_v1 | DMT400049096 | GCCAGTTAGCATCACAATCTGTATTGTCTTGTGTATATGTGTAATCTATAGGTTATGGAT |
| CUST_52483_PI426222305 | JHI_St_60k_v1 | DMT400049092 | TGAGTTTTTTCGTCAAACTCAGTCCGAGGAGATCTTTACAGGAATTGTGAGAGAAGTGCA |
| CUST_52482_PI426222305 | JHI_St_60k_v1 | DMT400049095 | GCCAGTTAGCATCACAATCTGTATTGTCTTGTGTATATGTGTAATCTATAGGTTATGGAT |
| CUST_52486_PI426222305 | JHI_St_60k_v1 | DMT400049094 | GCCAGTTAGCATCACAATCTGTATTGTCTTGTGTATATGTGTAATCTATAGGTTATGGAT |
Microarray Signals from CUST_52483_PI426222305
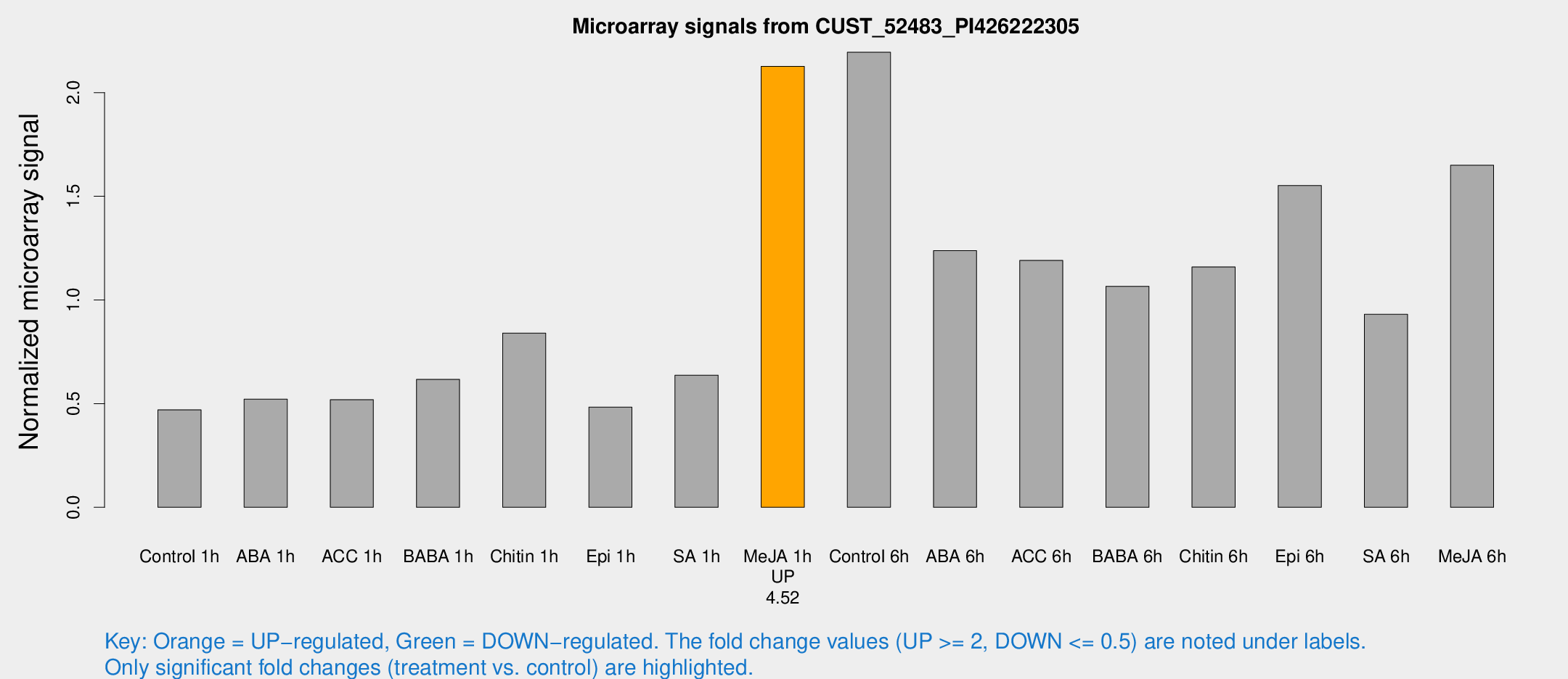
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 216.675 | 19.3225 | 0.470062 | 0.0279393 |
| ABA 1h | 221.54 | 50.6802 | 0.521708 | 0.0870096 |
| ACC 1h | 253.943 | 49.2269 | 0.519029 | 0.129499 |
| BABA 1h | 293.648 | 71.0905 | 0.617125 | 0.117922 |
| Chitin 1h | 347.114 | 22.8704 | 0.840077 | 0.0490366 |
| Epi 1h | 198.01 | 34.4926 | 0.483331 | 0.0837393 |
| SA 1h | 313.286 | 68.2405 | 0.637403 | 0.129794 |
| Me-JA 1h | 833.83 | 180.462 | 2.12635 | 0.293492 |
| Control 6h | 1069.02 | 249.718 | 2.19478 | 0.356671 |
| ABA 6h | 615.485 | 92.3824 | 1.23775 | 0.115388 |
| ACC 6h | 644.785 | 110.167 | 1.19075 | 0.110688 |
| BABA 6h | 547.716 | 32.3781 | 1.06588 | 0.0619099 |
| Chitin 6h | 565.185 | 32.8393 | 1.15952 | 0.0673113 |
| Epi 6h | 859.107 | 215.435 | 1.55174 | 0.540572 |
| SA 6h | 504.893 | 174.113 | 0.930911 | 0.358502 |
| Me-JA 6h | 771.834 | 118.45 | 1.64998 | 0.14327 |
Source Transcript PGSC0003DMT400049092 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G47240.1 | +1 | 2e-115 | 340 | 189/288 (66%) | nudix hydrolase homolog 8 | chr5:19183806-19185467 FORWARD LENGTH=369 |
