Probe CUST_52486_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_52486_PI426222305 | JHI_St_60k_v1 | DMT400049094 | GCCAGTTAGCATCACAATCTGTATTGTCTTGTGTATATGTGTAATCTATAGGTTATGGAT |
All Microarray Probes Designed to Gene DMG400019082
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_52484_PI426222305 | JHI_St_60k_v1 | DMT400049096 | GCCAGTTAGCATCACAATCTGTATTGTCTTGTGTATATGTGTAATCTATAGGTTATGGAT |
| CUST_52483_PI426222305 | JHI_St_60k_v1 | DMT400049092 | TGAGTTTTTTCGTCAAACTCAGTCCGAGGAGATCTTTACAGGAATTGTGAGAGAAGTGCA |
| CUST_52482_PI426222305 | JHI_St_60k_v1 | DMT400049095 | GCCAGTTAGCATCACAATCTGTATTGTCTTGTGTATATGTGTAATCTATAGGTTATGGAT |
| CUST_52486_PI426222305 | JHI_St_60k_v1 | DMT400049094 | GCCAGTTAGCATCACAATCTGTATTGTCTTGTGTATATGTGTAATCTATAGGTTATGGAT |
Microarray Signals from CUST_52486_PI426222305
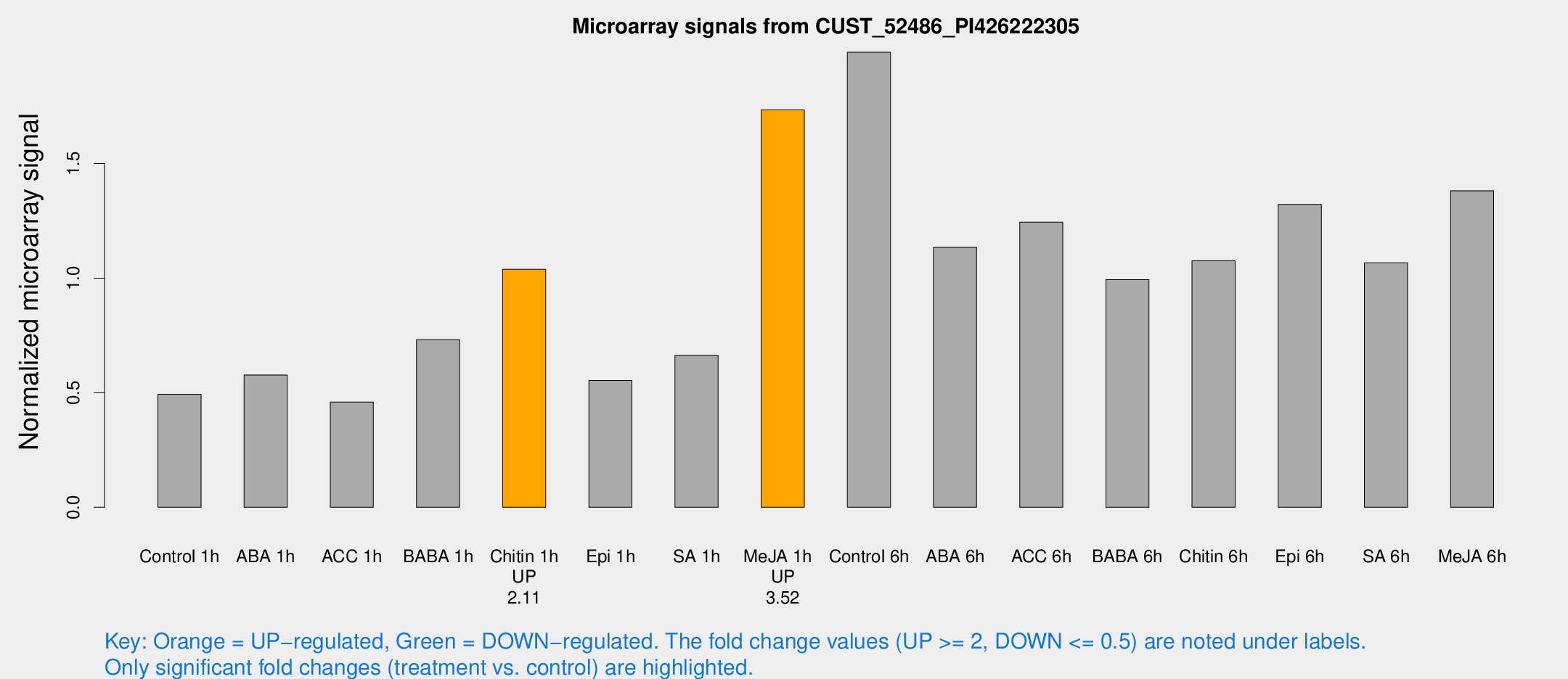
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 3808.81 | 429.43 | 0.493353 | 0.0545082 |
| ABA 1h | 3991.4 | 643.729 | 0.577587 | 0.0700317 |
| ACC 1h | 3667.36 | 518.564 | 0.459034 | 0.0857131 |
| BABA 1h | 5637.76 | 1151.94 | 0.731939 | 0.0926732 |
| Chitin 1h | 7123.82 | 411.688 | 1.03886 | 0.0623668 |
| Epi 1h | 3745.55 | 599.485 | 0.553271 | 0.078298 |
| SA 1h | 5304.73 | 816.398 | 0.662911 | 0.0989442 |
| Me-JA 1h | 11042.1 | 1607.85 | 1.73452 | 0.131864 |
| Control 6h | 15941.8 | 3492.29 | 1.9864 | 0.278262 |
| ABA 6h | 9432.13 | 1563.57 | 1.13488 | 0.120118 |
| ACC 6h | 11041.4 | 1321.53 | 1.24462 | 0.0822668 |
| BABA 6h | 8507.91 | 551.768 | 0.993527 | 0.060151 |
| Chitin 6h | 8753.19 | 507.042 | 1.07566 | 0.0974425 |
| Epi 6h | 11818 | 2208.78 | 1.32239 | 0.36179 |
| SA 6h | 8489.18 | 1809.37 | 1.06704 | 0.137839 |
| Me-JA 6h | 10948.5 | 2200.74 | 1.38198 | 0.176015 |
Source Transcript PGSC0003DMT400049094 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G47240.1 | +2 | 5e-71 | 212 | 117/175 (67%) | nudix hydrolase homolog 8 | chr5:19183806-19185467 FORWARD LENGTH=369 |
