Probe CUST_50625_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_50625_PI426222305 | JHI_St_60k_v1 | DMT400065527 | GCTAACTCATACAATATGTGATGTGTTGTTTCTACTAAACGAAAGCTTTGGATGTATGAC |
All Microarray Probes Designed to Gene DMG400025495
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_50625_PI426222305 | JHI_St_60k_v1 | DMT400065527 | GCTAACTCATACAATATGTGATGTGTTGTTTCTACTAAACGAAAGCTTTGGATGTATGAC |
| CUST_50635_PI426222305 | JHI_St_60k_v1 | DMT400065528 | ATTCTGTTTGGTCCTATGTTGCTCAGACTCTTCAAAAATGTCATCGGGTGCATGCCCCAT |
| CUST_50617_PI426222305 | JHI_St_60k_v1 | DMT400065526 | GCTAACTCATACAATATGTGATGTGTTGTTTCTACTAAACGAAAGCTTTGGATGTATGAC |
| CUST_50631_PI426222305 | JHI_St_60k_v1 | DMT400065525 | TGTGTGCTAACTCATACAATATGTGATGTGTTGTTTCTACTAAACGAAAGCTTTGGATGT |
Microarray Signals from CUST_50625_PI426222305
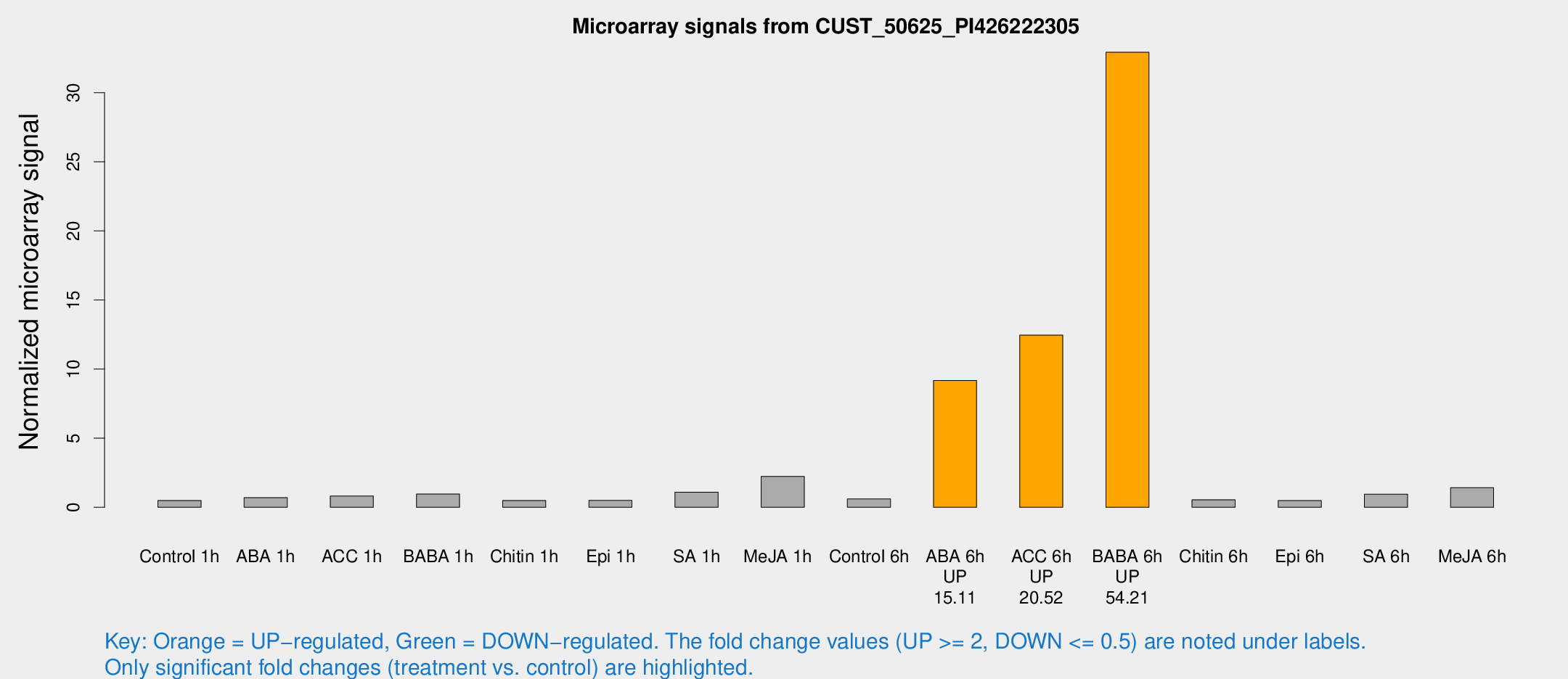
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 20.3128 | 3.46179 | 0.490331 | 0.0875902 |
| ABA 1h | 40.7647 | 23.4715 | 0.696513 | 1.07334 |
| ACC 1h | 36.5669 | 10.4282 | 0.816529 | 0.195022 |
| BABA 1h | 37.6636 | 4.05918 | 0.959874 | 0.153234 |
| Chitin 1h | 21.3232 | 7.41959 | 0.491312 | 0.287612 |
| Epi 1h | 18.0119 | 3.36888 | 0.496716 | 0.118429 |
| SA 1h | 68.9509 | 34.8248 | 1.09174 | 1.14867 |
| Me-JA 1h | 74.4275 | 8.09499 | 2.23709 | 0.24071 |
| Control 6h | 30.4729 | 11.0585 | 0.6074 | 0.297999 |
| ABA 6h | 446.178 | 146.48 | 9.17799 | 3.13937 |
| ACC 6h | 601.077 | 129.552 | 12.4612 | 3.90863 |
| BABA 6h | 1586.91 | 398.275 | 32.9298 | 8.77373 |
| Chitin 6h | 29.3891 | 11.2614 | 0.544963 | 0.357683 |
| Epi 6h | 28.7343 | 10.877 | 0.489552 | 0.305707 |
| SA 6h | 39.2269 | 7.7359 | 0.949644 | 0.132925 |
| Me-JA 6h | 62.2037 | 19.0321 | 1.42534 | 0.320004 |
Source Transcript PGSC0003DMT400065527 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G61800.1 | +1 | 2e-179 | 516 | 288/371 (78%) | glucose-6-phosphate/phosphate translocator 2 | chr1:22824527-22826459 FORWARD LENGTH=388 |
