Probe CUST_50617_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_50617_PI426222305 | JHI_St_60k_v1 | DMT400065526 | GCTAACTCATACAATATGTGATGTGTTGTTTCTACTAAACGAAAGCTTTGGATGTATGAC |
All Microarray Probes Designed to Gene DMG400025495
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_50625_PI426222305 | JHI_St_60k_v1 | DMT400065527 | GCTAACTCATACAATATGTGATGTGTTGTTTCTACTAAACGAAAGCTTTGGATGTATGAC |
| CUST_50635_PI426222305 | JHI_St_60k_v1 | DMT400065528 | ATTCTGTTTGGTCCTATGTTGCTCAGACTCTTCAAAAATGTCATCGGGTGCATGCCCCAT |
| CUST_50617_PI426222305 | JHI_St_60k_v1 | DMT400065526 | GCTAACTCATACAATATGTGATGTGTTGTTTCTACTAAACGAAAGCTTTGGATGTATGAC |
| CUST_50631_PI426222305 | JHI_St_60k_v1 | DMT400065525 | TGTGTGCTAACTCATACAATATGTGATGTGTTGTTTCTACTAAACGAAAGCTTTGGATGT |
Microarray Signals from CUST_50617_PI426222305
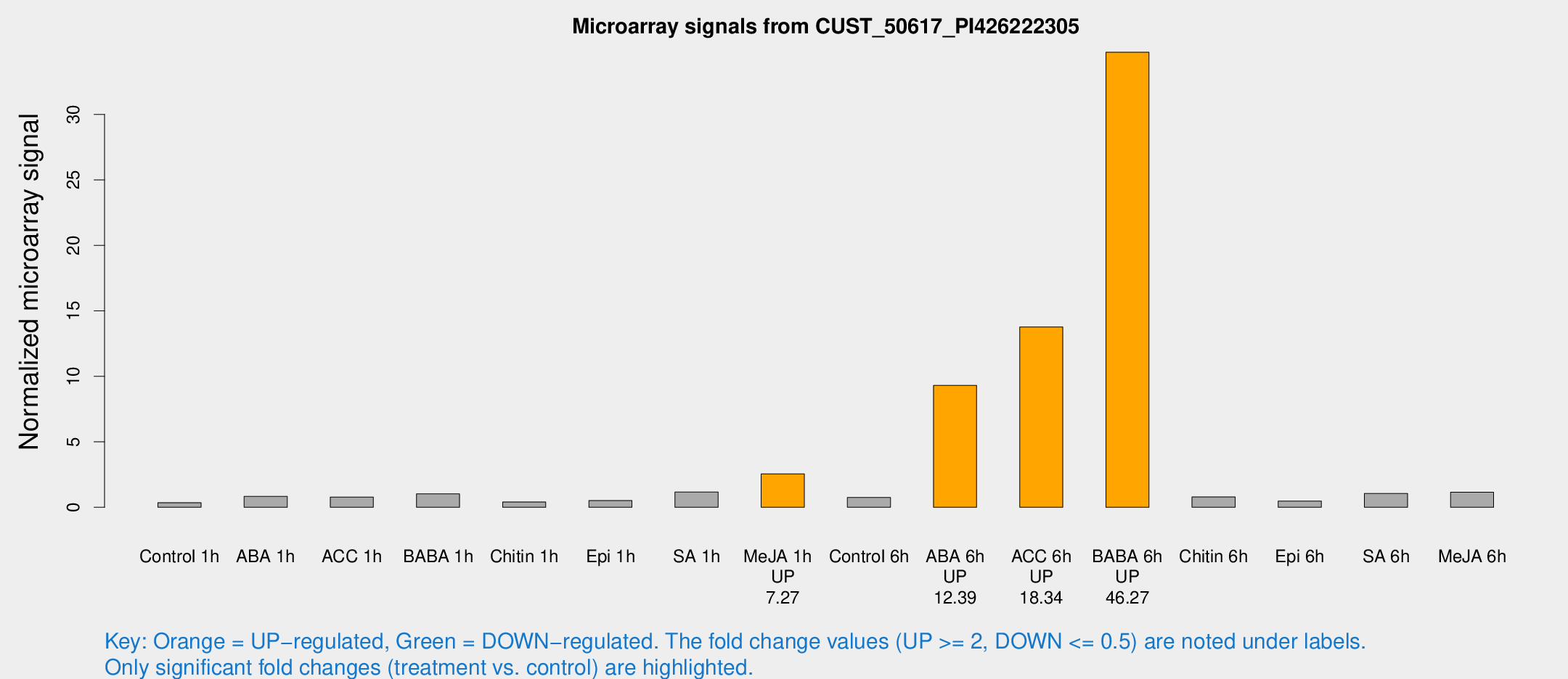
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 16.7606 | 6.10309 | 0.351383 | 0.175448 |
| ABA 1h | 36.832 | 18.0101 | 0.829788 | 0.566275 |
| ACC 1h | 33.6417 | 9.16765 | 0.781863 | 0.175924 |
| BABA 1h | 38.8973 | 4.11553 | 1.02691 | 0.109407 |
| Chitin 1h | 15.1266 | 3.35435 | 0.406835 | 0.0983908 |
| Epi 1h | 18.1403 | 3.71331 | 0.514531 | 0.108556 |
| SA 1h | 62.5891 | 30.0463 | 1.16109 | 0.803207 |
| Me-JA 1h | 82.9011 | 10.5605 | 2.5555 | 0.179508 |
| Control 6h | 30.3443 | 5.30143 | 0.750873 | 0.0998251 |
| ABA 6h | 440.464 | 138.866 | 9.3046 | 3.40717 |
| ACC 6h | 637.694 | 118.973 | 13.7709 | 3.78171 |
| BABA 6h | 1674.66 | 489.185 | 34.7423 | 12.0125 |
| Chitin 6h | 34.4799 | 7.79648 | 0.790713 | 0.157363 |
| Epi 6h | 28.2686 | 13.6335 | 0.475211 | 0.298963 |
| SA 6h | 42.974 | 9.12927 | 1.05372 | 0.347666 |
| Me-JA 6h | 53.0811 | 21.1696 | 1.1471 | 0.429332 |
Source Transcript PGSC0003DMT400065526 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G61800.1 | +3 | 2e-72 | 233 | 134/150 (89%) | glucose-6-phosphate/phosphate translocator 2 | chr1:22824527-22826459 FORWARD LENGTH=388 |
