Probe CUST_50426_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_50426_PI426222305 | JHI_St_60k_v1 | DMT400010912 | AACTTGCGGATTATATTCAAAACAGAAATCATGCAATCAAGATGGCTGCTCGGTTTCTCC |
All Microarray Probes Designed to Gene DMG400004274
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_50398_PI426222305 | JHI_St_60k_v1 | DMT400010911 | AACTTGCGGATTATATTCAAAACAGAAATCATGCAATCAAGATGGCTGCTCGGTTTCTCC |
| CUST_50411_PI426222305 | JHI_St_60k_v1 | DMT400010913 | TTCCATAGTGCGAGGAGGCCGAAGATTCCCCTTGTATAGGTAAGTGACTAATGTTATGCT |
| CUST_50426_PI426222305 | JHI_St_60k_v1 | DMT400010912 | AACTTGCGGATTATATTCAAAACAGAAATCATGCAATCAAGATGGCTGCTCGGTTTCTCC |
Microarray Signals from CUST_50426_PI426222305
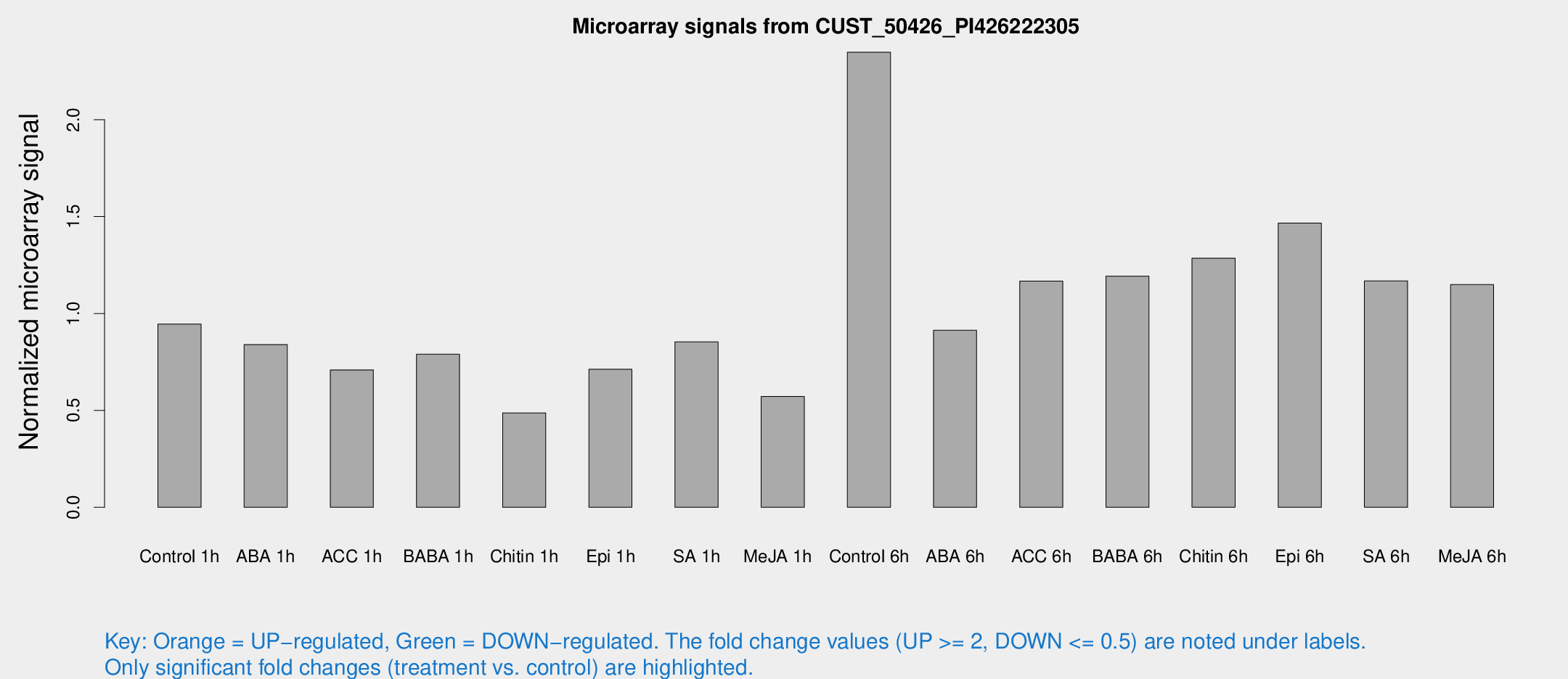
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 126.932 | 34.7135 | 0.945513 | 0.191015 |
| ABA 1h | 93.8877 | 9.57351 | 0.839585 | 0.0789952 |
| ACC 1h | 111.896 | 40.8006 | 0.709115 | 0.343791 |
| BABA 1h | 102.289 | 26.5098 | 0.78989 | 0.147205 |
| Chitin 1h | 55.2337 | 4.99646 | 0.487135 | 0.0408081 |
| Epi 1h | 77.2019 | 5.70008 | 0.712092 | 0.0526653 |
| SA 1h | 109.947 | 7.29068 | 0.853334 | 0.0565062 |
| Me-JA 1h | 58.5006 | 4.98559 | 0.572041 | 0.0486645 |
| Control 6h | 305.653 | 59.2719 | 2.34805 | 0.327904 |
| ABA 6h | 121.683 | 8.01079 | 0.913489 | 0.085461 |
| ACC 6h | 169.865 | 19.5743 | 1.1669 | 0.141294 |
| BABA 6h | 172.21 | 30.8784 | 1.1927 | 0.190652 |
| Chitin 6h | 172.28 | 13.4556 | 1.28548 | 0.101055 |
| Epi 6h | 207.447 | 12.7535 | 1.46663 | 0.0898565 |
| SA 6h | 151.931 | 32.3373 | 1.16769 | 0.339493 |
| Me-JA 6h | 148.978 | 30.8735 | 1.149 | 0.14313 |
Source Transcript PGSC0003DMT400010912 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G55320.1 | +1 | 0.0 | 807 | 406/635 (64%) | acyl-activating enzyme 18 | chr1:20633371-20636659 FORWARD LENGTH=727 |
