Probe CUST_50398_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_50398_PI426222305 | JHI_St_60k_v1 | DMT400010911 | AACTTGCGGATTATATTCAAAACAGAAATCATGCAATCAAGATGGCTGCTCGGTTTCTCC |
All Microarray Probes Designed to Gene DMG400004274
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_50398_PI426222305 | JHI_St_60k_v1 | DMT400010911 | AACTTGCGGATTATATTCAAAACAGAAATCATGCAATCAAGATGGCTGCTCGGTTTCTCC |
| CUST_50411_PI426222305 | JHI_St_60k_v1 | DMT400010913 | TTCCATAGTGCGAGGAGGCCGAAGATTCCCCTTGTATAGGTAAGTGACTAATGTTATGCT |
| CUST_50426_PI426222305 | JHI_St_60k_v1 | DMT400010912 | AACTTGCGGATTATATTCAAAACAGAAATCATGCAATCAAGATGGCTGCTCGGTTTCTCC |
Microarray Signals from CUST_50398_PI426222305
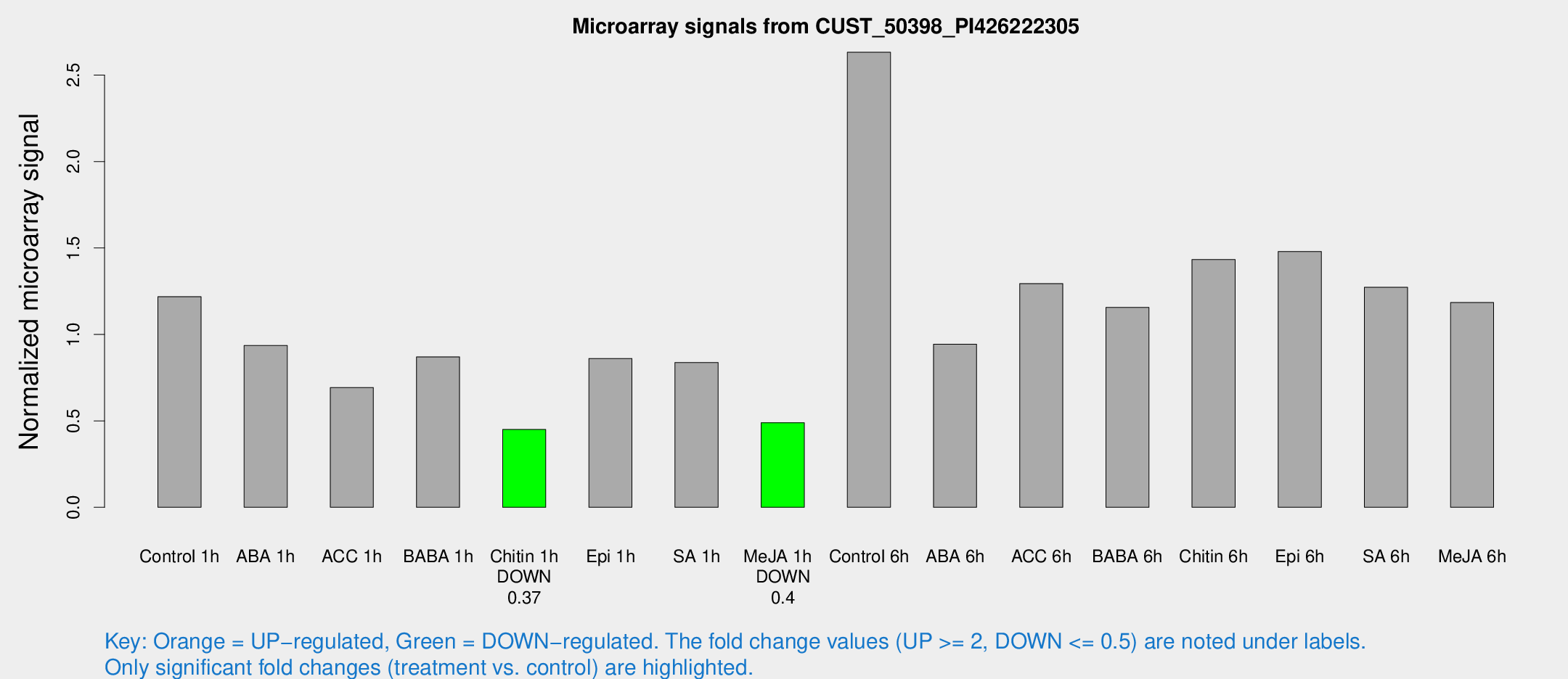
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 149.117 | 36.8782 | 1.2185 | 0.230019 |
| ABA 1h | 97.3019 | 11.8415 | 0.935722 | 0.0897346 |
| ACC 1h | 93.2153 | 27.9357 | 0.692973 | 0.225273 |
| BABA 1h | 98.7677 | 12.5033 | 0.870069 | 0.0619296 |
| Chitin 1h | 47.0651 | 4.64805 | 0.450002 | 0.044558 |
| Epi 1h | 86.8482 | 7.25384 | 0.860947 | 0.0625111 |
| SA 1h | 101.371 | 13.7194 | 0.837143 | 0.152869 |
| Me-JA 1h | 46.2985 | 4.65497 | 0.489365 | 0.0491226 |
| Control 6h | 314.474 | 51.4488 | 2.63242 | 0.25275 |
| ABA 6h | 116.572 | 8.35022 | 0.943044 | 0.0728442 |
| ACC 6h | 174.117 | 19.4514 | 1.29414 | 0.196985 |
| BABA 6h | 152.794 | 22.656 | 1.15599 | 0.158708 |
| Chitin 6h | 176.911 | 11.0671 | 1.43319 | 0.08954 |
| Epi 6h | 198.767 | 35.2942 | 1.47993 | 0.251236 |
| SA 6h | 153.377 | 36.6357 | 1.27253 | 0.235843 |
| Me-JA 6h | 139.928 | 23.0187 | 1.18444 | 0.132892 |
Source Transcript PGSC0003DMT400010911 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G55320.1 | +1 | 0.0 | 828 | 406/591 (69%) | acyl-activating enzyme 18 | chr1:20633371-20636659 FORWARD LENGTH=727 |
