Probe CUST_5034_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_5034_PI426222305 | JHI_St_60k_v1 | DMT400003939 | GTTGTTGTTGAAATCTATTATTGTTTCCAGTCATGTTCTTGGAGCATATGCTAGTTTCCT |
All Microarray Probes Designed to Gene DMG400001551
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_5181_PI426222305 | JHI_St_60k_v1 | DMT400003940 | GTTCTTAAAGAAGATCCATCTAACCCTTTGTTTTTGAGGAATTATGCTCAACTTTTGCAG |
| CUST_5428_PI426222305 | JHI_St_60k_v1 | DMT400003938 | GTTCTTAAAGAAGATCCATCTAACCCTTTGTTTTTGAGGAATTATGCTCAACTTTTGCAG |
| CUST_5034_PI426222305 | JHI_St_60k_v1 | DMT400003939 | GTTGTTGTTGAAATCTATTATTGTTTCCAGTCATGTTCTTGGAGCATATGCTAGTTTCCT |
| CUST_5324_PI426222305 | JHI_St_60k_v1 | DMT400003937 | AGATAAAGCCTCAACCTACTTTAAGAGGGCAGTTCAAGCTTCTCCAGAAGACGGAAAAAA |
Microarray Signals from CUST_5034_PI426222305
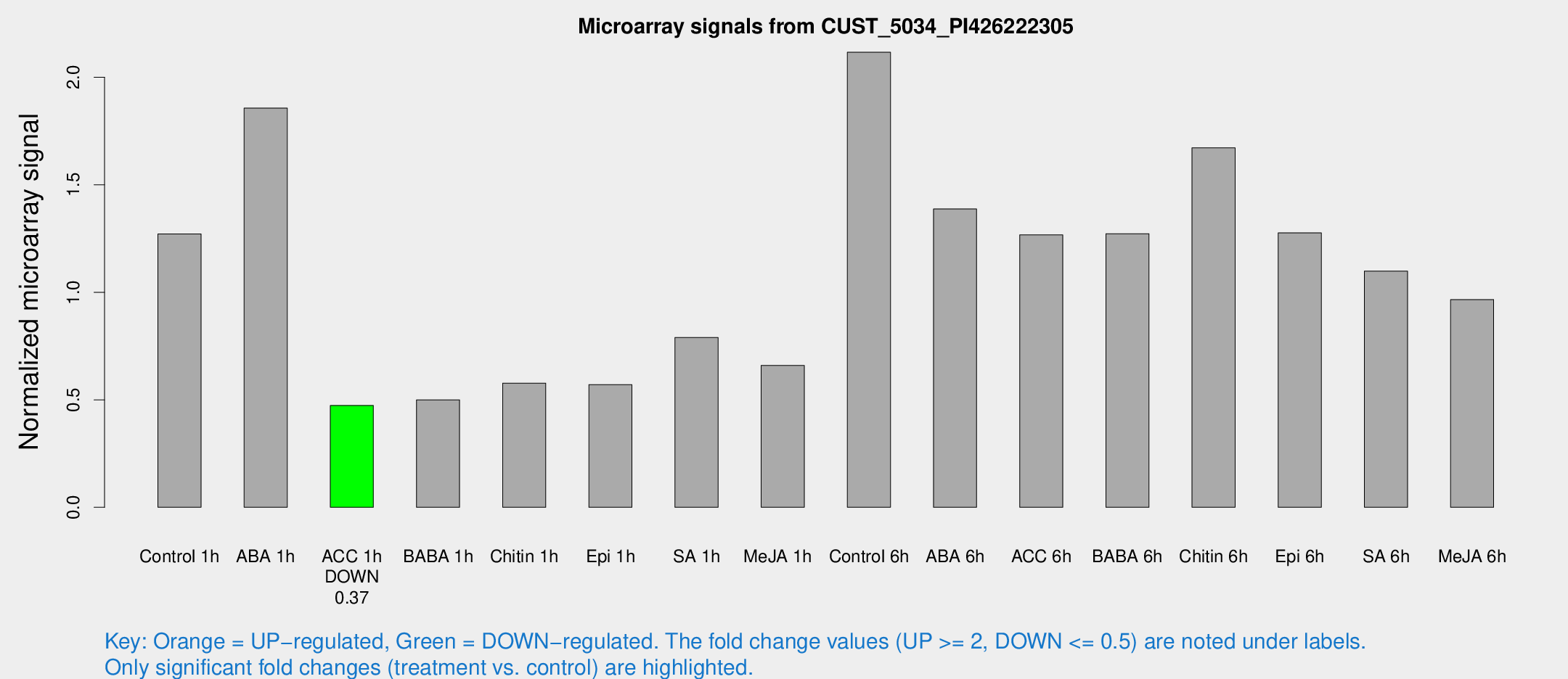
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 18.4156 | 4.54516 | 1.27134 | 0.312742 |
| ABA 1h | 22.4158 | 3.30884 | 1.85672 | 0.277433 |
| ACC 1h | 6.63062 | 3.41765 | 0.473691 | 0.246111 |
| BABA 1h | 6.59965 | 3.33058 | 0.499681 | 0.254863 |
| Chitin 1h | 7.44026 | 3.2127 | 0.577421 | 0.276433 |
| Epi 1h | 6.74516 | 3.13335 | 0.570472 | 0.269512 |
| SA 1h | 12.9154 | 4.87309 | 0.790028 | 0.344009 |
| Me-JA 1h | 8.21025 | 3.28509 | 0.659747 | 0.31727 |
| Control 6h | 32.084 | 9.86409 | 2.11651 | 0.585804 |
| ABA 6h | 21.3568 | 4.82538 | 1.38736 | 0.323525 |
| ACC 6h | 20.5816 | 4.1853 | 1.26678 | 0.28069 |
| BABA 6h | 22.6936 | 9.49359 | 1.27259 | 0.532719 |
| Chitin 6h | 24.3375 | 3.9136 | 1.67171 | 0.272456 |
| Epi 6h | 22.5735 | 8.08287 | 1.27641 | 0.486132 |
| SA 6h | 15.4207 | 3.62388 | 1.09888 | 0.28858 |
| Me-JA 6h | 15.2669 | 4.94121 | 0.965965 | 0.384554 |
Source Transcript PGSC0003DMT400003939 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G04530.1 | +2 | 8e-24 | 101 | 72/181 (40%) | Tetratricopeptide repeat (TPR)-like superfamily protein | chr1:1234456-1235895 REVERSE LENGTH=310 |
