Probe CUST_5181_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_5181_PI426222305 | JHI_St_60k_v1 | DMT400003940 | GTTCTTAAAGAAGATCCATCTAACCCTTTGTTTTTGAGGAATTATGCTCAACTTTTGCAG |
All Microarray Probes Designed to Gene DMG400001551
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_5181_PI426222305 | JHI_St_60k_v1 | DMT400003940 | GTTCTTAAAGAAGATCCATCTAACCCTTTGTTTTTGAGGAATTATGCTCAACTTTTGCAG |
| CUST_5428_PI426222305 | JHI_St_60k_v1 | DMT400003938 | GTTCTTAAAGAAGATCCATCTAACCCTTTGTTTTTGAGGAATTATGCTCAACTTTTGCAG |
| CUST_5034_PI426222305 | JHI_St_60k_v1 | DMT400003939 | GTTGTTGTTGAAATCTATTATTGTTTCCAGTCATGTTCTTGGAGCATATGCTAGTTTCCT |
| CUST_5324_PI426222305 | JHI_St_60k_v1 | DMT400003937 | AGATAAAGCCTCAACCTACTTTAAGAGGGCAGTTCAAGCTTCTCCAGAAGACGGAAAAAA |
Microarray Signals from CUST_5181_PI426222305
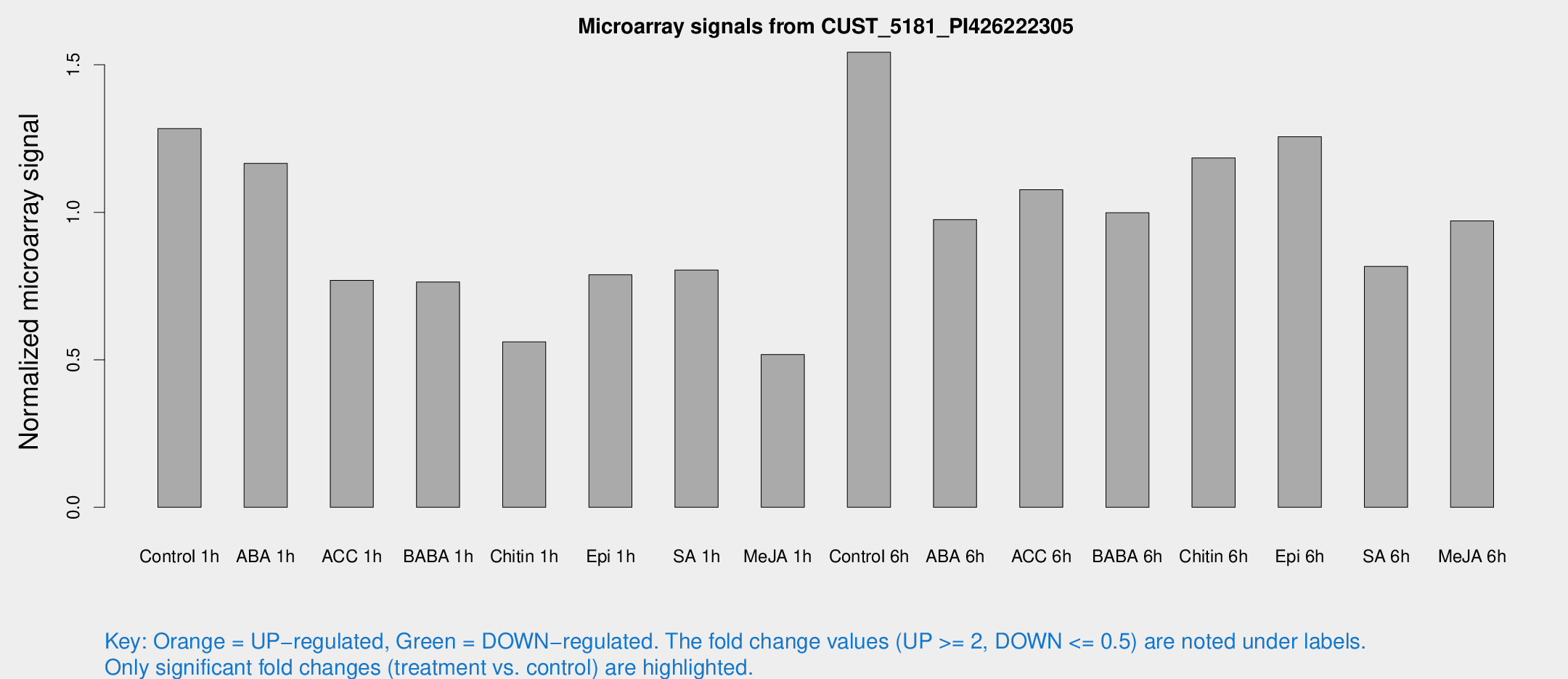
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 179.688 | 26.9739 | 1.28409 | 0.106047 |
| ABA 1h | 141.365 | 8.90289 | 1.16608 | 0.0733819 |
| ACC 1h | 113.288 | 22.5089 | 0.76955 | 0.116742 |
| BABA 1h | 111.551 | 30.5571 | 0.76384 | 0.187584 |
| Chitin 1h | 73.3732 | 18.7726 | 0.560623 | 0.113612 |
| Epi 1h | 93.883 | 7.27449 | 0.788014 | 0.0723101 |
| SA 1h | 122.271 | 31.9342 | 0.80414 | 0.190819 |
| Me-JA 1h | 62.7727 | 18.4802 | 0.518023 | 0.104424 |
| Control 6h | 232.847 | 65.1568 | 1.54262 | 0.38707 |
| ABA 6h | 150.167 | 32.4489 | 0.975424 | 0.212496 |
| ACC 6h | 175.182 | 32.1914 | 1.07673 | 0.100518 |
| BABA 6h | 158.425 | 29.5619 | 0.998487 | 0.167453 |
| Chitin 6h | 175.118 | 21.6966 | 1.18453 | 0.117934 |
| Epi 6h | 202.971 | 43.0373 | 1.25645 | 0.232809 |
| SA 6h | 122.438 | 34.2578 | 0.816791 | 0.189438 |
| Me-JA 6h | 152.462 | 52.0974 | 0.971092 | 0.323225 |
Source Transcript PGSC0003DMT400003940 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G04530.1 | +3 | 1e-36 | 139 | 114/341 (33%) | Tetratricopeptide repeat (TPR)-like superfamily protein | chr1:1234456-1235895 REVERSE LENGTH=310 |
