Probe CUST_46616_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_46616_PI426222305 | JHI_St_60k_v1 | DMT400027036 | GTAAGCGATACAAATTCCACTATTTTAGTTTAGGTCCTAAATACGTGTGCTTACATCAGT |
All Microarray Probes Designed to Gene DMG400010430
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_46627_PI426222305 | JHI_St_60k_v1 | DMT400027035 | TTAAGATCGGGAAAGCTCAAAAGACAGAACCACCTTCATAATTATAATGTTTGTGCATGA |
| CUST_46616_PI426222305 | JHI_St_60k_v1 | DMT400027036 | GTAAGCGATACAAATTCCACTATTTTAGTTTAGGTCCTAAATACGTGTGCTTACATCAGT |
| CUST_46635_PI426222305 | JHI_St_60k_v1 | DMT400027037 | GTAAGCGATACAAATTCCACTATTTTAGTTTAGGTCCTAAATACGTGTGCTTACATCAGT |
Microarray Signals from CUST_46616_PI426222305
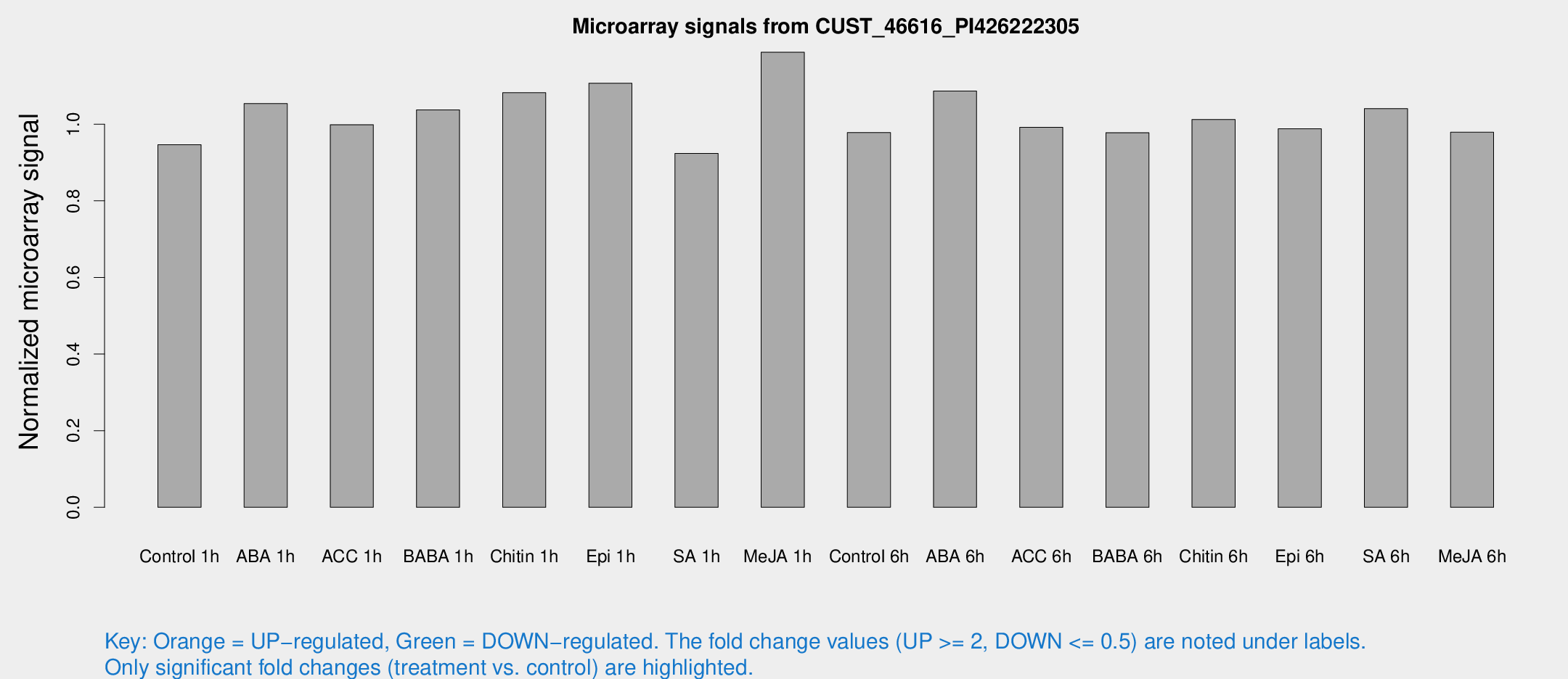
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 5.2005 | 3.01295 | 0.946299 | 0.548038 |
| ABA 1h | 5.00311 | 2.90158 | 1.05384 | 0.595842 |
| ACC 1h | 5.64184 | 3.27127 | 0.998725 | 0.578336 |
| BABA 1h | 5.49555 | 3.18361 | 1.03738 | 0.600699 |
| Chitin 1h | 5.22931 | 3.03878 | 1.08211 | 0.611212 |
| Epi 1h | 5.22748 | 3.03253 | 1.10694 | 0.635541 |
| SA 1h | 5.21371 | 3.01952 | 0.923754 | 0.534931 |
| Me-JA 1h | 5.31681 | 3.0861 | 1.18787 | 0.685093 |
| Control 6h | 5.38684 | 3.11978 | 0.978262 | 0.566507 |
| ABA 6h | 6.48489 | 3.25571 | 1.0864 | 0.565394 |
| ACC 6h | 6.4087 | 3.80945 | 0.991951 | 0.574411 |
| BABA 6h | 6.02398 | 3.49493 | 0.977776 | 0.566211 |
| Chitin 6h | 5.92206 | 3.42999 | 1.01238 | 0.586195 |
| Epi 6h | 6.1661 | 3.59802 | 0.98799 | 0.572102 |
| SA 6h | 5.659 | 3.27891 | 1.0408 | 0.602708 |
| Me-JA 6h | 5.35942 | 3.1101 | 0.97895 | 0.567773 |
Source Transcript PGSC0003DMT400027036 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G54070.1 | +1 | 3e-77 | 265 | 199/658 (30%) | Ankyrin repeat family protein | chr3:20021330-20023603 REVERSE LENGTH=574 |
