Probe CUST_46635_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_46635_PI426222305 | JHI_St_60k_v1 | DMT400027037 | GTAAGCGATACAAATTCCACTATTTTAGTTTAGGTCCTAAATACGTGTGCTTACATCAGT |
All Microarray Probes Designed to Gene DMG400010430
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_46627_PI426222305 | JHI_St_60k_v1 | DMT400027035 | TTAAGATCGGGAAAGCTCAAAAGACAGAACCACCTTCATAATTATAATGTTTGTGCATGA |
| CUST_46616_PI426222305 | JHI_St_60k_v1 | DMT400027036 | GTAAGCGATACAAATTCCACTATTTTAGTTTAGGTCCTAAATACGTGTGCTTACATCAGT |
| CUST_46635_PI426222305 | JHI_St_60k_v1 | DMT400027037 | GTAAGCGATACAAATTCCACTATTTTAGTTTAGGTCCTAAATACGTGTGCTTACATCAGT |
Microarray Signals from CUST_46635_PI426222305
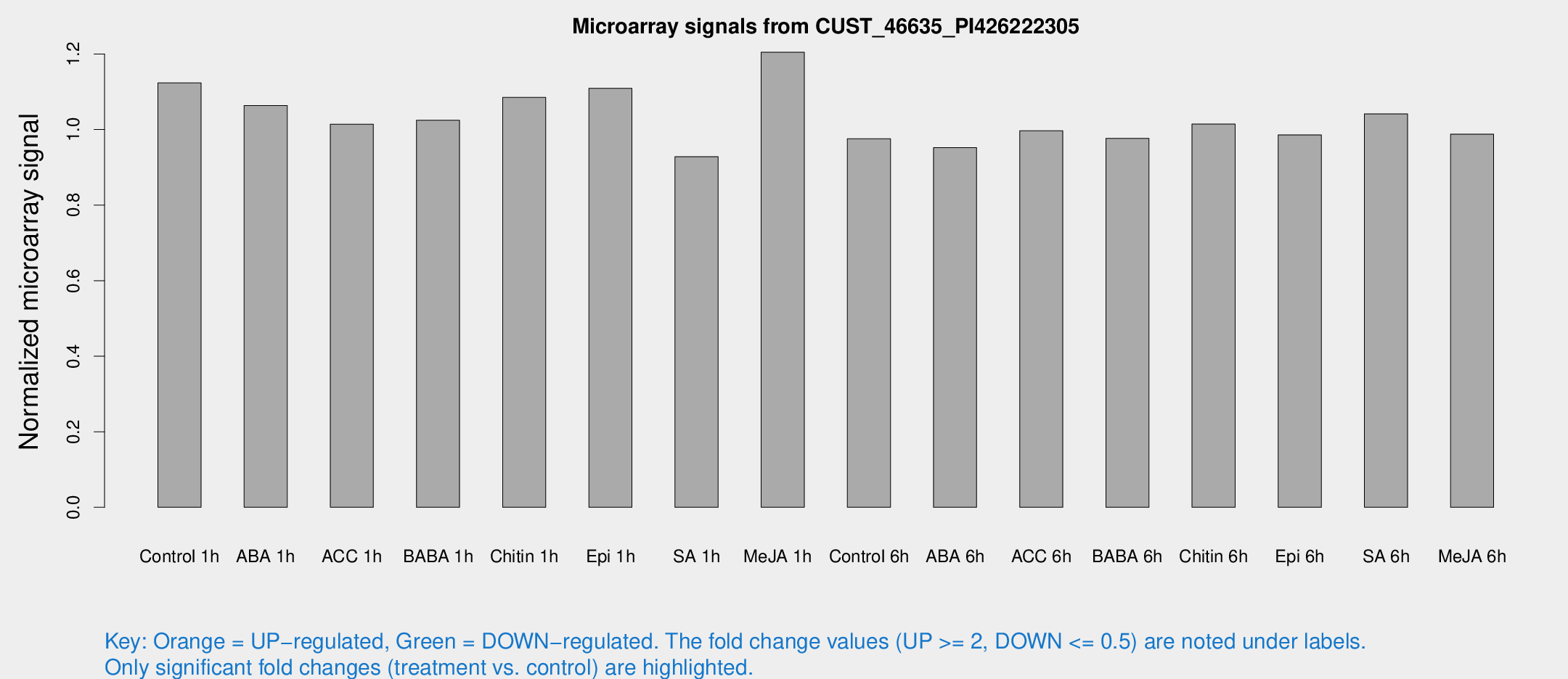
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 6.13111 | 2.91016 | 1.12358 | 0.564319 |
| ABA 1h | 4.8433 | 2.81036 | 1.06335 | 0.595552 |
| ACC 1h | 5.5671 | 3.23184 | 1.01429 | 0.587349 |
| BABA 1h | 5.27437 | 3.05957 | 1.02457 | 0.593297 |
| Chitin 1h | 5.0202 | 2.92065 | 1.08519 | 0.604497 |
| Epi 1h | 4.97013 | 2.88366 | 1.1093 | 0.623081 |
| SA 1h | 5.03294 | 2.91664 | 0.927986 | 0.532023 |
| Me-JA 1h | 5.14121 | 2.98794 | 1.20473 | 0.681563 |
| Control 6h | 5.2137 | 3.01984 | 0.975488 | 0.5649 |
| ABA 6h | 5.40202 | 3.13082 | 0.952175 | 0.551366 |
| ACC 6h | 6.22326 | 3.68275 | 0.996741 | 0.577183 |
| BABA 6h | 5.84171 | 3.39079 | 0.976571 | 0.565514 |
| Chitin 6h | 5.76018 | 3.33654 | 1.01455 | 0.587452 |
| Epi 6h | 5.97322 | 3.48685 | 0.985832 | 0.570872 |
| SA 6h | 5.4947 | 3.18349 | 1.04136 | 0.603033 |
| Me-JA 6h | 5.21947 | 3.02673 | 0.987907 | 0.569148 |
Source Transcript PGSC0003DMT400027037 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G35810.1 | +2 | 9e-54 | 192 | 101/204 (50%) | Ankyrin repeat family protein | chr5:13993428-13994549 REVERSE LENGTH=347 |
