Probe CUST_46135_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_46135_PI426222305 | JHI_St_60k_v1 | DMT400018474 | TTGACAGGATCTTGCTTACCTACCTTAAAAACAGAACAGAAACTTCTAGCAAGTCCCATA |
All Microarray Probes Designed to Gene DMG400007169
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_46157_PI426222305 | JHI_St_60k_v1 | DMT400018473 | TCCTTGTATGTACCCTTACCTACCTTAAAAACAGAACAGAAACTTCTAGCAAGTCCCATA |
| CUST_46110_PI426222305 | JHI_St_60k_v1 | DMT400018472 | CGTGGTGCTATTGCTTATAGTTCCTCTAAGGCTGCTATGGATTCTATGACGTTCATGAAC |
| CUST_46135_PI426222305 | JHI_St_60k_v1 | DMT400018474 | TTGACAGGATCTTGCTTACCTACCTTAAAAACAGAACAGAAACTTCTAGCAAGTCCCATA |
Microarray Signals from CUST_46135_PI426222305
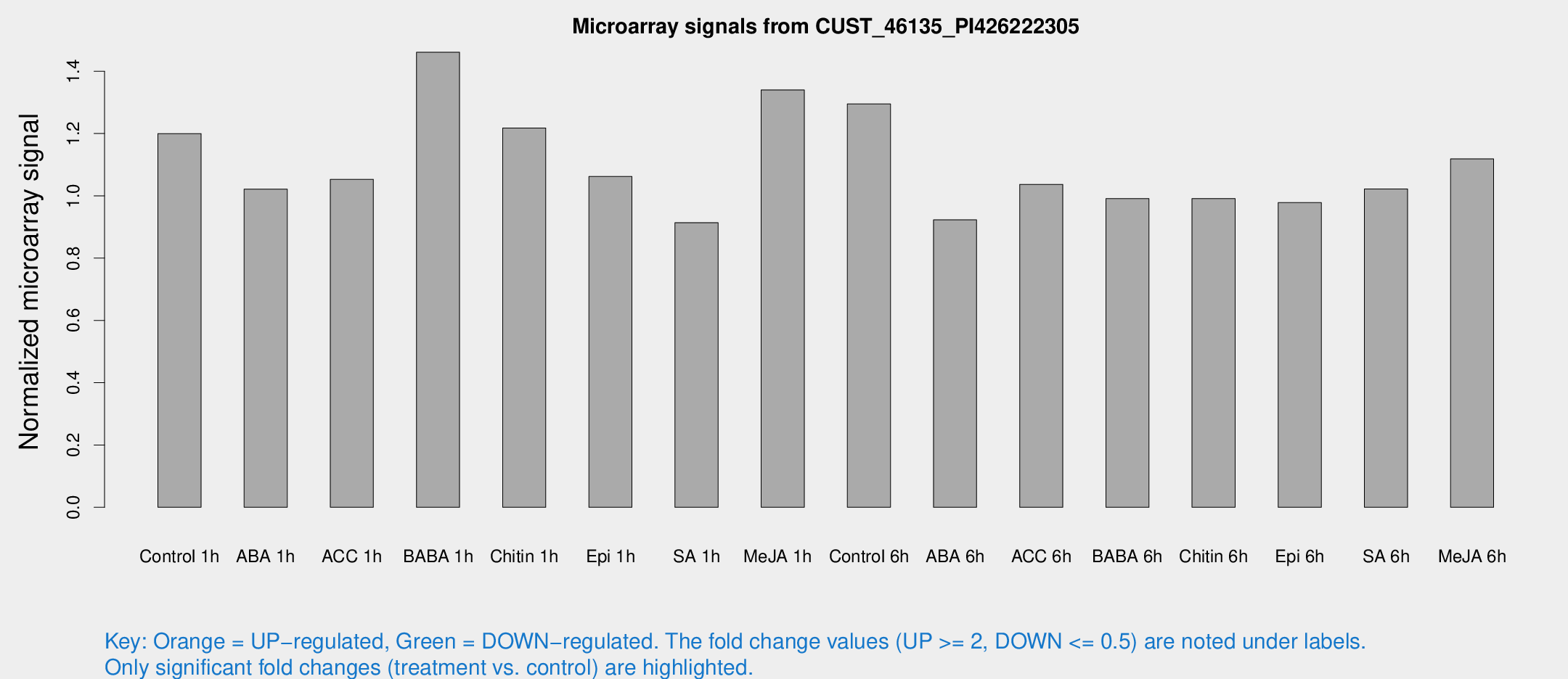
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 7.16791 | 3.0932 | 1.19995 | 0.561962 |
| ABA 1h | 5.12853 | 2.97458 | 1.02168 | 0.585109 |
| ACC 1h | 6.30759 | 3.72031 | 1.0528 | 0.609875 |
| BABA 1h | 9.1672 | 3.42582 | 1.4611 | 0.68192 |
| Chitin 1h | 6.34791 | 3.07483 | 1.218 | 0.606269 |
| Epi 1h | 5.20949 | 3.02401 | 1.06197 | 0.607152 |
| SA 1h | 5.387 | 3.12435 | 0.913837 | 0.529816 |
| Me-JA 1h | 6.36448 | 3.19172 | 1.34001 | 0.691494 |
| Control 6h | 8.25719 | 3.1792 | 1.29527 | 0.607839 |
| ABA 6h | 5.63335 | 3.26558 | 0.922907 | 0.534513 |
| ACC 6h | 6.93557 | 3.89145 | 1.03684 | 0.566688 |
| BABA 6h | 6.41154 | 3.54979 | 0.991113 | 0.551618 |
| Chitin 6h | 6.05356 | 3.51045 | 0.991284 | 0.574796 |
| Epi 6h | 6.37319 | 3.72015 | 0.978147 | 0.56658 |
| SA 6h | 5.80351 | 3.36332 | 1.02207 | 0.59186 |
| Me-JA 6h | 6.52228 | 3.1857 | 1.11852 | 0.565251 |
Source Transcript PGSC0003DMT400018474 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G55290.1 | +1 | 6e-49 | 168 | 78/125 (62%) | NAD(P)-binding Rossmann-fold superfamily protein | chr3:20502653-20503730 FORWARD LENGTH=280 |
