Probe CUST_46110_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_46110_PI426222305 | JHI_St_60k_v1 | DMT400018472 | CGTGGTGCTATTGCTTATAGTTCCTCTAAGGCTGCTATGGATTCTATGACGTTCATGAAC |
All Microarray Probes Designed to Gene DMG400007169
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_46157_PI426222305 | JHI_St_60k_v1 | DMT400018473 | TCCTTGTATGTACCCTTACCTACCTTAAAAACAGAACAGAAACTTCTAGCAAGTCCCATA |
| CUST_46110_PI426222305 | JHI_St_60k_v1 | DMT400018472 | CGTGGTGCTATTGCTTATAGTTCCTCTAAGGCTGCTATGGATTCTATGACGTTCATGAAC |
| CUST_46135_PI426222305 | JHI_St_60k_v1 | DMT400018474 | TTGACAGGATCTTGCTTACCTACCTTAAAAACAGAACAGAAACTTCTAGCAAGTCCCATA |
Microarray Signals from CUST_46110_PI426222305
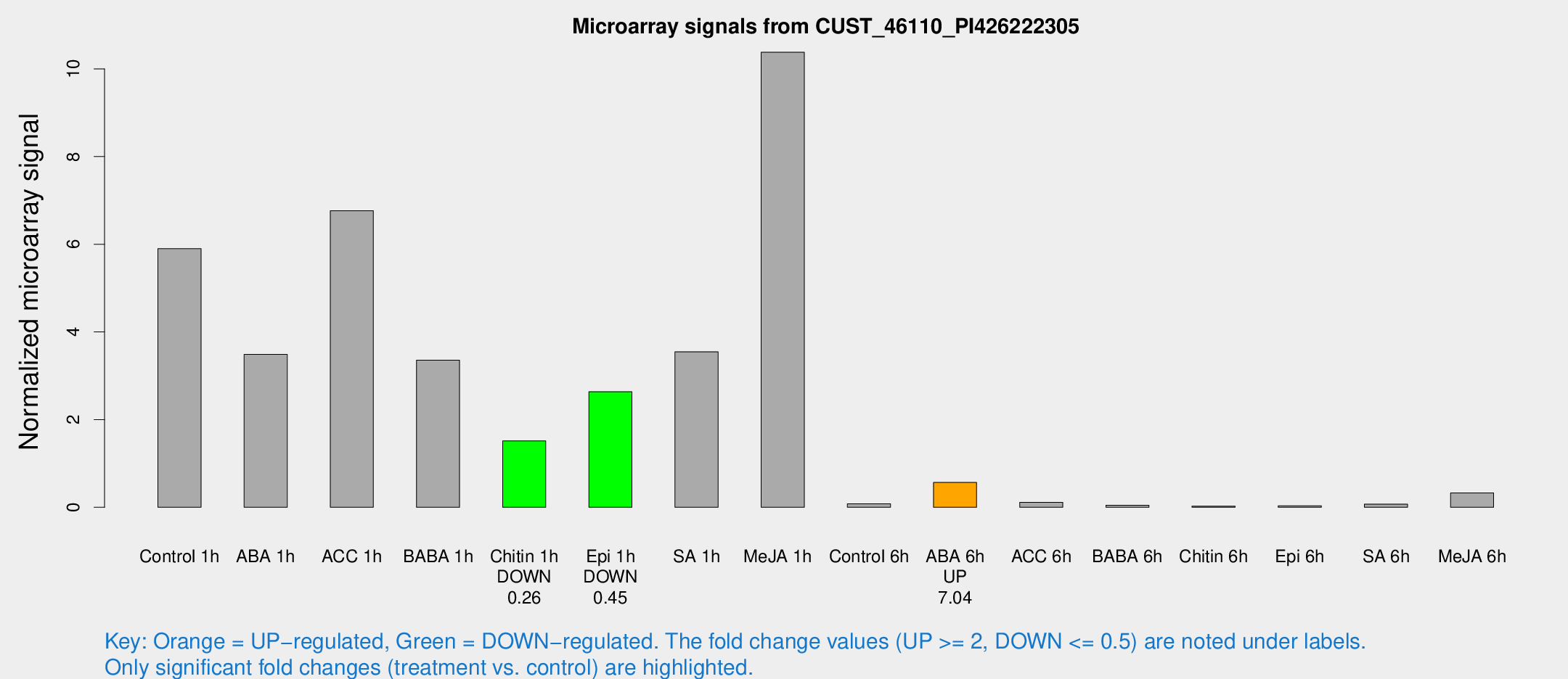
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 4122.88 | 875.837 | 5.90038 | 0.827564 |
| ABA 1h | 2373 | 733.494 | 3.48907 | 1.3118 |
| ACC 1h | 5628.87 | 2202.47 | 6.76512 | 2.99502 |
| BABA 1h | 2270.7 | 498.927 | 3.35602 | 0.50429 |
| Chitin 1h | 954.258 | 185.005 | 1.51453 | 0.251529 |
| Epi 1h | 1547.51 | 178.278 | 2.63617 | 0.318874 |
| SA 1h | 2990.87 | 1361.3 | 3.54696 | 1.96788 |
| Me-JA 1h | 5891.82 | 1143.11 | 10.3807 | 1.10621 |
| Control 6h | 61.9631 | 23.6113 | 0.0806008 | 0.0248084 |
| ABA 6h | 413.654 | 63.8698 | 0.567614 | 0.0692538 |
| ACC 6h | 99.2347 | 35.8792 | 0.11474 | 0.0584638 |
| BABA 6h | 56.4239 | 38.2389 | 0.0466522 | 0.0429914 |
| Chitin 6h | 22.5667 | 7.35964 | 0.0270604 | 0.0146747 |
| Epi 6h | 25.6992 | 4.37323 | 0.034003 | 0.00583839 |
| SA 6h | 48.0098 | 4.86341 | 0.0717587 | 0.0161493 |
| Me-JA 6h | 264.579 | 119.249 | 0.329367 | 0.1969 |
Source Transcript PGSC0003DMT400018472 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G55290.1 | +1 | 6e-111 | 325 | 169/270 (63%) | NAD(P)-binding Rossmann-fold superfamily protein | chr3:20502653-20503730 FORWARD LENGTH=280 |
