Probe CUST_46098_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_46098_PI426222305 | JHI_St_60k_v1 | DMT400018544 | CACCACATTGTGAAATAATGGTCACGAGTTTATATAGTATAACTATGGCGATGTAATGTC |
All Microarray Probes Designed to Gene DMG400007196
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_46086_PI426222305 | JHI_St_60k_v1 | DMT400018546 | GATTTCCTCGGCGACGGTATTTTTAACGTTGATGGTGAAATTTGGAAAATTCAAAGGAAA |
| CUST_46097_PI426222305 | JHI_St_60k_v1 | DMT400018545 | TCTGCTAGCTATGAATTTAGTACCAGATCTCTAAGGAATTTCGTTATGGAAACTGCACAG |
| CUST_46098_PI426222305 | JHI_St_60k_v1 | DMT400018544 | CACCACATTGTGAAATAATGGTCACGAGTTTATATAGTATAACTATGGCGATGTAATGTC |
Microarray Signals from CUST_46098_PI426222305
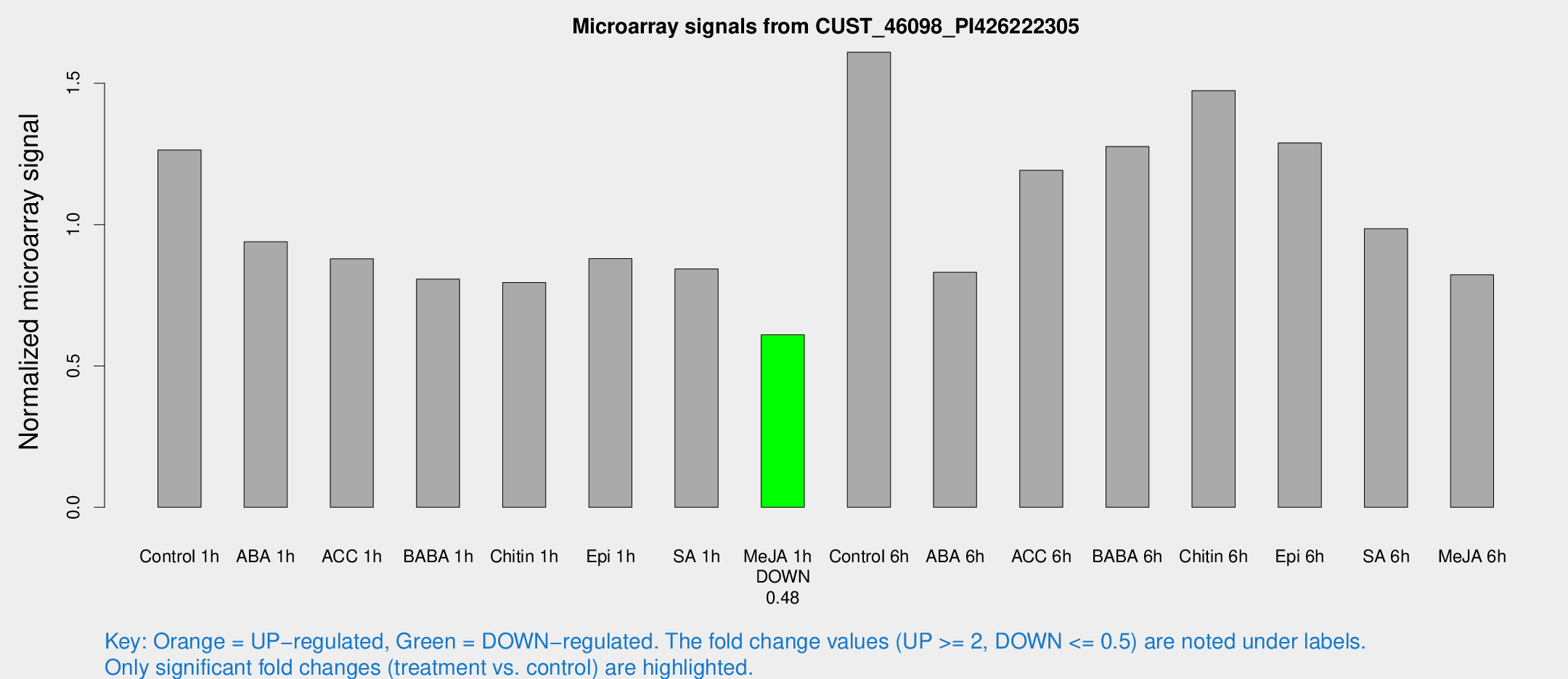
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 1372.95 | 142.38 | 1.26408 | 0.073044 |
| ABA 1h | 917.121 | 151.118 | 0.939424 | 0.0873338 |
| ACC 1h | 1015.96 | 197.842 | 0.879287 | 0.128867 |
| BABA 1h | 899.111 | 214.996 | 0.807146 | 0.147828 |
| Chitin 1h | 770.886 | 44.7712 | 0.794951 | 0.0460143 |
| Epi 1h | 828.779 | 90.3289 | 0.880041 | 0.0803123 |
| SA 1h | 944.419 | 115.862 | 0.843372 | 0.0540359 |
| Me-JA 1h | 545.142 | 75.6235 | 0.610164 | 0.0354261 |
| Control 6h | 1810.53 | 361.91 | 1.60983 | 0.225421 |
| ABA 6h | 955.207 | 71.0685 | 0.8316 | 0.0481058 |
| ACC 6h | 1539.92 | 307.166 | 1.19195 | 0.182391 |
| BABA 6h | 1582.61 | 285.463 | 1.27621 | 0.207473 |
| Chitin 6h | 1716.34 | 237.773 | 1.47364 | 0.157495 |
| Epi 6h | 1568.34 | 101.873 | 1.28904 | 0.123321 |
| SA 6h | 1189.88 | 355.007 | 0.985307 | 0.277822 |
| Me-JA 6h | 969.552 | 277.67 | 0.822433 | 0.204884 |
Source Transcript PGSC0003DMT400018544 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G56630.1 | +2 | 1e-144 | 437 | 218/352 (62%) | cytochrome P450, family 94, subfamily D, polypeptide 2 | chr3:20978953-20980512 FORWARD LENGTH=499 |
