Probe CUST_46097_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_46097_PI426222305 | JHI_St_60k_v1 | DMT400018545 | TCTGCTAGCTATGAATTTAGTACCAGATCTCTAAGGAATTTCGTTATGGAAACTGCACAG |
All Microarray Probes Designed to Gene DMG400007196
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_46086_PI426222305 | JHI_St_60k_v1 | DMT400018546 | GATTTCCTCGGCGACGGTATTTTTAACGTTGATGGTGAAATTTGGAAAATTCAAAGGAAA |
| CUST_46097_PI426222305 | JHI_St_60k_v1 | DMT400018545 | TCTGCTAGCTATGAATTTAGTACCAGATCTCTAAGGAATTTCGTTATGGAAACTGCACAG |
| CUST_46098_PI426222305 | JHI_St_60k_v1 | DMT400018544 | CACCACATTGTGAAATAATGGTCACGAGTTTATATAGTATAACTATGGCGATGTAATGTC |
Microarray Signals from CUST_46097_PI426222305
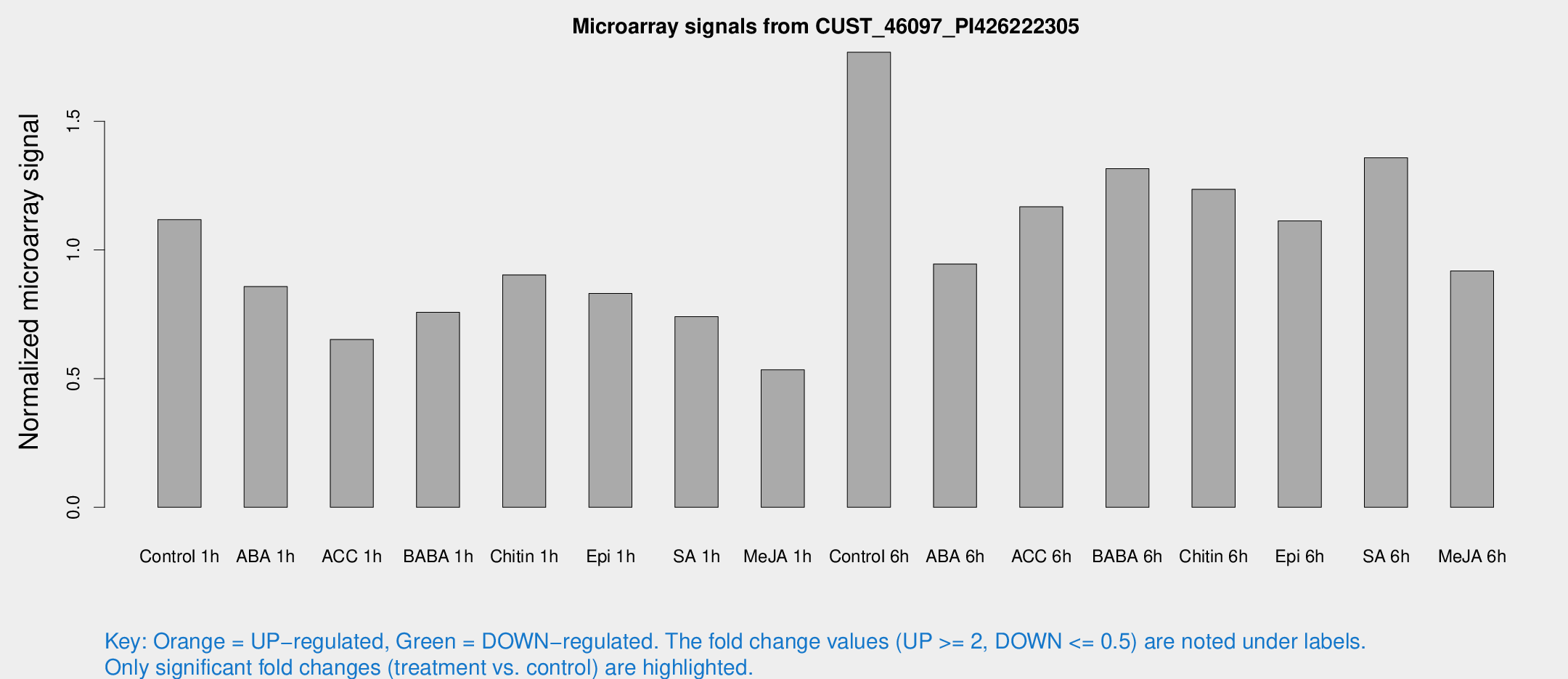
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 171.043 | 12.9805 | 1.11753 | 0.0683081 |
| ABA 1h | 119.979 | 21.7236 | 0.857547 | 0.129733 |
| ACC 1h | 122.736 | 42.3646 | 0.652319 | 0.292755 |
| BABA 1h | 123.054 | 34.1214 | 0.75741 | 0.186819 |
| Chitin 1h | 126.675 | 19.1162 | 0.903244 | 0.110205 |
| Epi 1h | 111.766 | 15.759 | 0.830757 | 0.107957 |
| SA 1h | 121.875 | 24.8962 | 0.741042 | 0.138429 |
| Me-JA 1h | 71.0988 | 18.8412 | 0.53412 | 0.122911 |
| Control 6h | 291.73 | 79.0308 | 1.76823 | 0.392109 |
| ABA 6h | 160.453 | 31.8773 | 0.945537 | 0.180132 |
| ACC 6h | 227.473 | 64.6611 | 1.16759 | 0.340953 |
| BABA 6h | 233.595 | 45.6841 | 1.31569 | 0.240958 |
| Chitin 6h | 210.339 | 43.2675 | 1.23584 | 0.290055 |
| Epi 6h | 201.035 | 46.28 | 1.11321 | 0.299442 |
| SA 6h | 222.743 | 58.3331 | 1.35811 | 0.619151 |
| Me-JA 6h | 153.718 | 44.6407 | 0.91808 | 0.234854 |
Source Transcript PGSC0003DMT400018545 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G56630.1 | +3 | 0.0 | 597 | 300/501 (60%) | cytochrome P450, family 94, subfamily D, polypeptide 2 | chr3:20978953-20980512 FORWARD LENGTH=499 |
