Probe CUST_45749_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_45749_PI426222305 | JHI_St_60k_v1 | DMT400055640 | TTTCAGTACTCTGGTTTATTGGCTTTGTTGTGGGGTATGAGAGACATACCCTGCACCGTA |
All Microarray Probes Designed to Gene DMG400021607
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_45743_PI426222305 | JHI_St_60k_v1 | DMT400055641 | CACCTTGAGATTAAGCTTTGTTTCTGGTTATATACAATCTCTCTGATGTACCATTTTGCT |
| CUST_45735_PI426222305 | JHI_St_60k_v1 | DMT400055643 | CTGGATATACTGTGTAACCCTGTCCTAACTTCTAGATGCGATAAATACTTATCTCAGGTG |
| CUST_45749_PI426222305 | JHI_St_60k_v1 | DMT400055640 | TTTCAGTACTCTGGTTTATTGGCTTTGTTGTGGGGTATGAGAGACATACCCTGCACCGTA |
| CUST_45759_PI426222305 | JHI_St_60k_v1 | DMT400055642 | CACCTTGAGATTAAGCTTTGTTTCTGGTTATATACAATCTCTCTGATGTACCATTTTGCT |
| CUST_45762_PI426222305 | JHI_St_60k_v1 | DMT400055645 | CACCTTGAGATTAAGCTTTGTTTCTGGTTATATACAATCTCTCTGATGTACCATTTTGCT |
Microarray Signals from CUST_45749_PI426222305
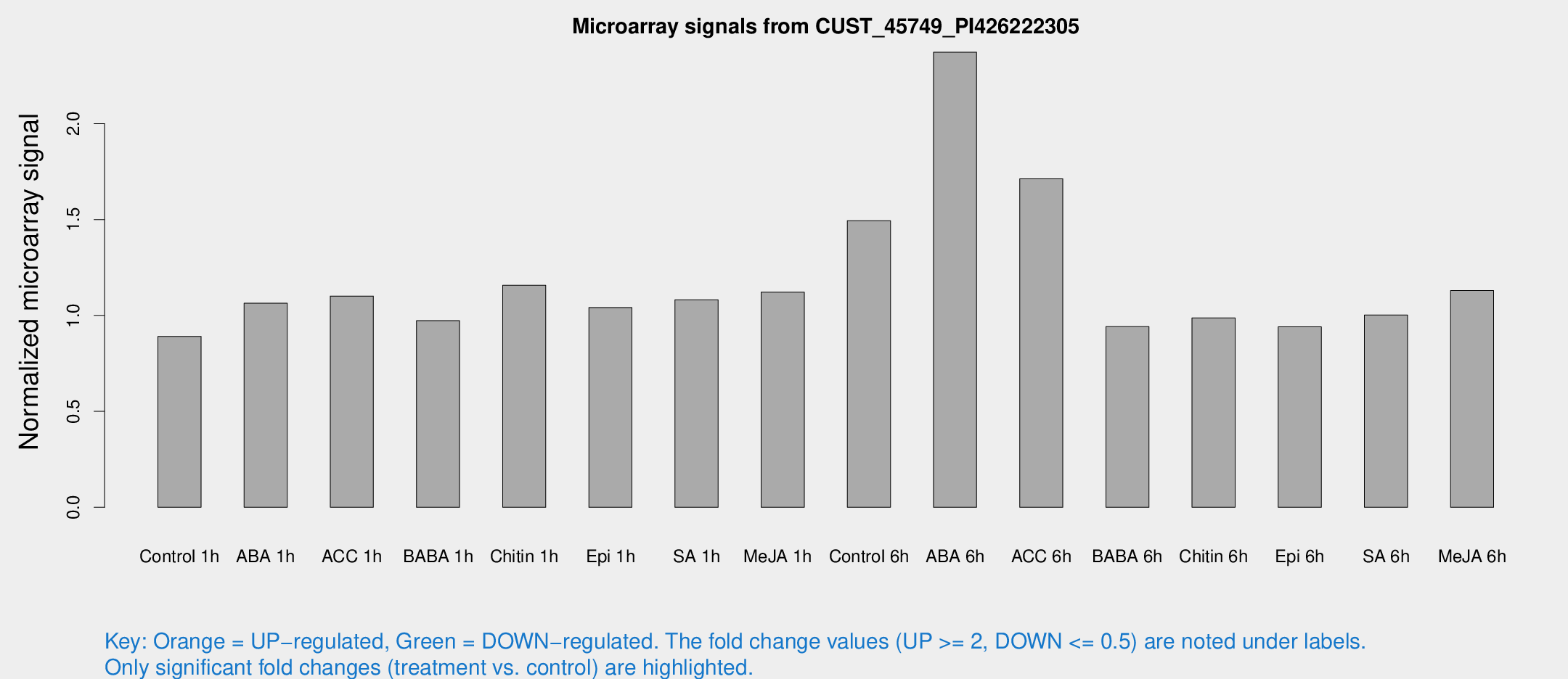
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 5.3341 | 3.09298 | 0.891218 | 0.514768 |
| ABA 1h | 5.67074 | 3.02052 | 1.06403 | 0.573065 |
| ACC 1h | 6.9144 | 3.41154 | 1.10107 | 0.566137 |
| BABA 1h | 5.63176 | 3.2703 | 0.97266 | 0.563462 |
| Chitin 1h | 6.38744 | 3.17282 | 1.15727 | 0.589841 |
| Epi 1h | 5.34087 | 3.07791 | 1.04133 | 0.59065 |
| SA 1h | 7.15532 | 3.10833 | 1.08175 | 0.53096 |
| Me-JA 1h | 5.51379 | 3.20767 | 1.12189 | 0.649592 |
| Control 6h | 9.70938 | 3.37362 | 1.49437 | 0.620169 |
| ABA 6h | 15.4712 | 3.51111 | 2.37212 | 0.566637 |
| ACC 6h | 15.6252 | 8.42638 | 1.71245 | 1.34887 |
| BABA 6h | 6.33788 | 3.68649 | 0.941807 | 0.545908 |
| Chitin 6h | 6.2999 | 3.61725 | 0.987134 | 0.566477 |
| Epi 6h | 6.42326 | 3.76357 | 0.940805 | 0.545534 |
| SA 6h | 5.94181 | 3.44125 | 1.00224 | 0.580378 |
| Me-JA 6h | 7.03457 | 3.32209 | 1.12966 | 0.571895 |
Source Transcript PGSC0003DMT400055640 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G56190.2 | +2 | 3e-51 | 177 | 83/128 (65%) | Transducin/WD40 repeat-like superfamily protein | chr5:22742654-22744909 FORWARD LENGTH=447 |
