Probe CUST_45735_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_45735_PI426222305 | JHI_St_60k_v1 | DMT400055643 | CTGGATATACTGTGTAACCCTGTCCTAACTTCTAGATGCGATAAATACTTATCTCAGGTG |
All Microarray Probes Designed to Gene DMG400021607
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_45743_PI426222305 | JHI_St_60k_v1 | DMT400055641 | CACCTTGAGATTAAGCTTTGTTTCTGGTTATATACAATCTCTCTGATGTACCATTTTGCT |
| CUST_45735_PI426222305 | JHI_St_60k_v1 | DMT400055643 | CTGGATATACTGTGTAACCCTGTCCTAACTTCTAGATGCGATAAATACTTATCTCAGGTG |
| CUST_45749_PI426222305 | JHI_St_60k_v1 | DMT400055640 | TTTCAGTACTCTGGTTTATTGGCTTTGTTGTGGGGTATGAGAGACATACCCTGCACCGTA |
| CUST_45759_PI426222305 | JHI_St_60k_v1 | DMT400055642 | CACCTTGAGATTAAGCTTTGTTTCTGGTTATATACAATCTCTCTGATGTACCATTTTGCT |
| CUST_45762_PI426222305 | JHI_St_60k_v1 | DMT400055645 | CACCTTGAGATTAAGCTTTGTTTCTGGTTATATACAATCTCTCTGATGTACCATTTTGCT |
Microarray Signals from CUST_45735_PI426222305
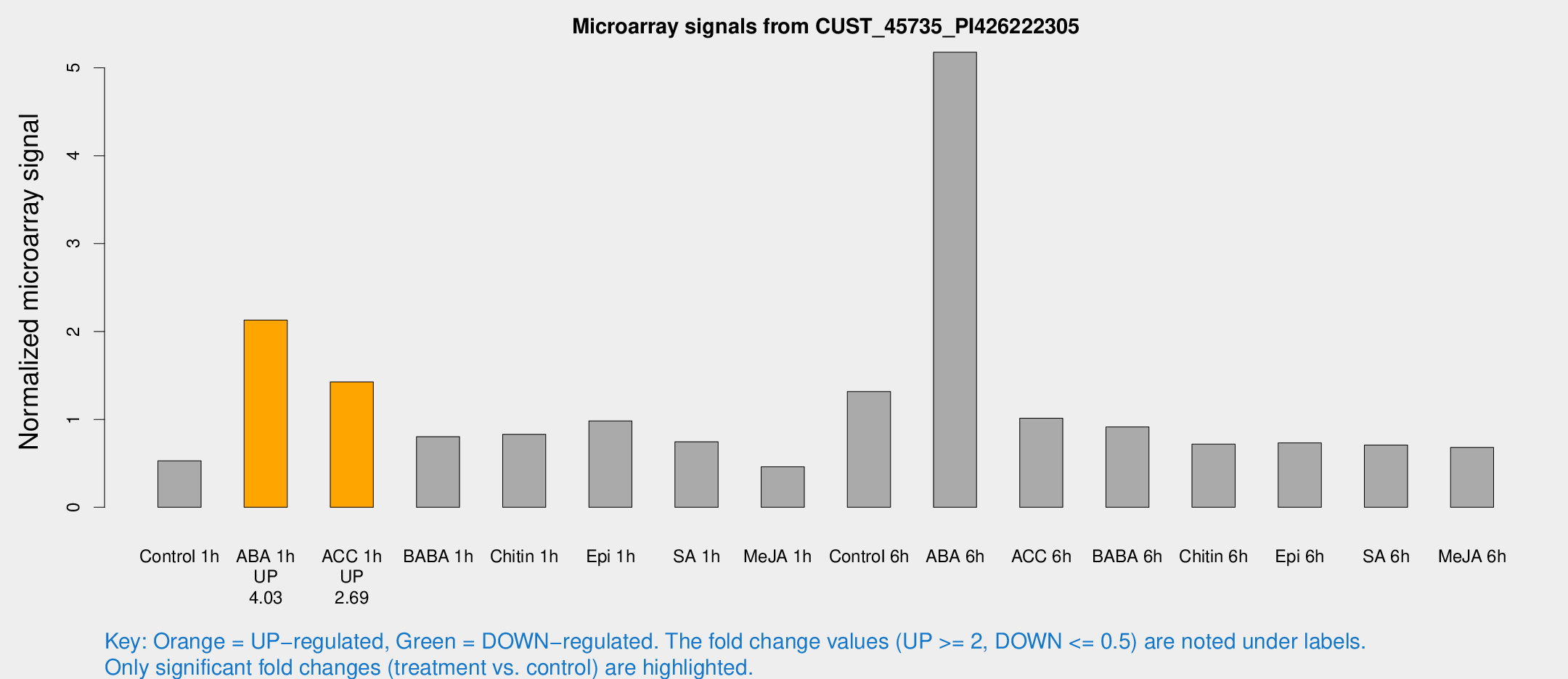
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 8.85971 | 3.51194 | 0.529193 | 0.225851 |
| ABA 1h | 31.3116 | 5.08465 | 2.13034 | 0.299324 |
| ACC 1h | 25.0369 | 5.39645 | 1.42595 | 0.356621 |
| BABA 1h | 13.968 | 4.14267 | 0.80341 | 0.284952 |
| Chitin 1h | 12.1253 | 3.48952 | 0.830913 | 0.24369 |
| Epi 1h | 15.6948 | 4.75622 | 0.983171 | 0.421235 |
| SA 1h | 12.4566 | 3.51433 | 0.745448 | 0.214819 |
| Me-JA 1h | 6.08009 | 3.5291 | 0.460551 | 0.266775 |
| Control 6h | 21.4375 | 3.749 | 1.31651 | 0.234932 |
| ABA 6h | 98.2977 | 27.9445 | 5.17711 | 1.56355 |
| ACC 6h | 18.9943 | 4.47375 | 1.01295 | 0.266958 |
| BABA 6h | 18.7219 | 5.57974 | 0.913845 | 0.348084 |
| Chitin 6h | 14.0879 | 5.15479 | 0.718229 | 0.30675 |
| Epi 6h | 17.0444 | 8.74975 | 0.733412 | 0.414517 |
| SA 6h | 16.061 | 9.66306 | 0.708609 | 0.730748 |
| Me-JA 6h | 11.9744 | 3.63847 | 0.680497 | 0.256952 |
Source Transcript PGSC0003DMT400055643 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G13340.1 | +2 | 8e-32 | 121 | 54/84 (64%) | Transducin/WD40 repeat-like superfamily protein | chr3:4332370-4334603 FORWARD LENGTH=447 |
