Probe CUST_39961_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_39961_PI426222305 | JHI_St_60k_v1 | DMT400081473 | GCTTATGCTAATCATTTTCTTGAAGGATCGTTTGGTAGAGTGCATTAGAAAAGATAATGC |
All Microarray Probes Designed to Gene DMG400031868
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_39961_PI426222305 | JHI_St_60k_v1 | DMT400081473 | GCTTATGCTAATCATTTTCTTGAAGGATCGTTTGGTAGAGTGCATTAGAAAAGATAATGC |
| CUST_39962_PI426222305 | JHI_St_60k_v1 | DMT400081474 | GCTTATGCTAATCATTTTCTTGAAGGATCGTTTGGTAGAGTGCATTAGAAAAGATAATGC |
Microarray Signals from CUST_39961_PI426222305
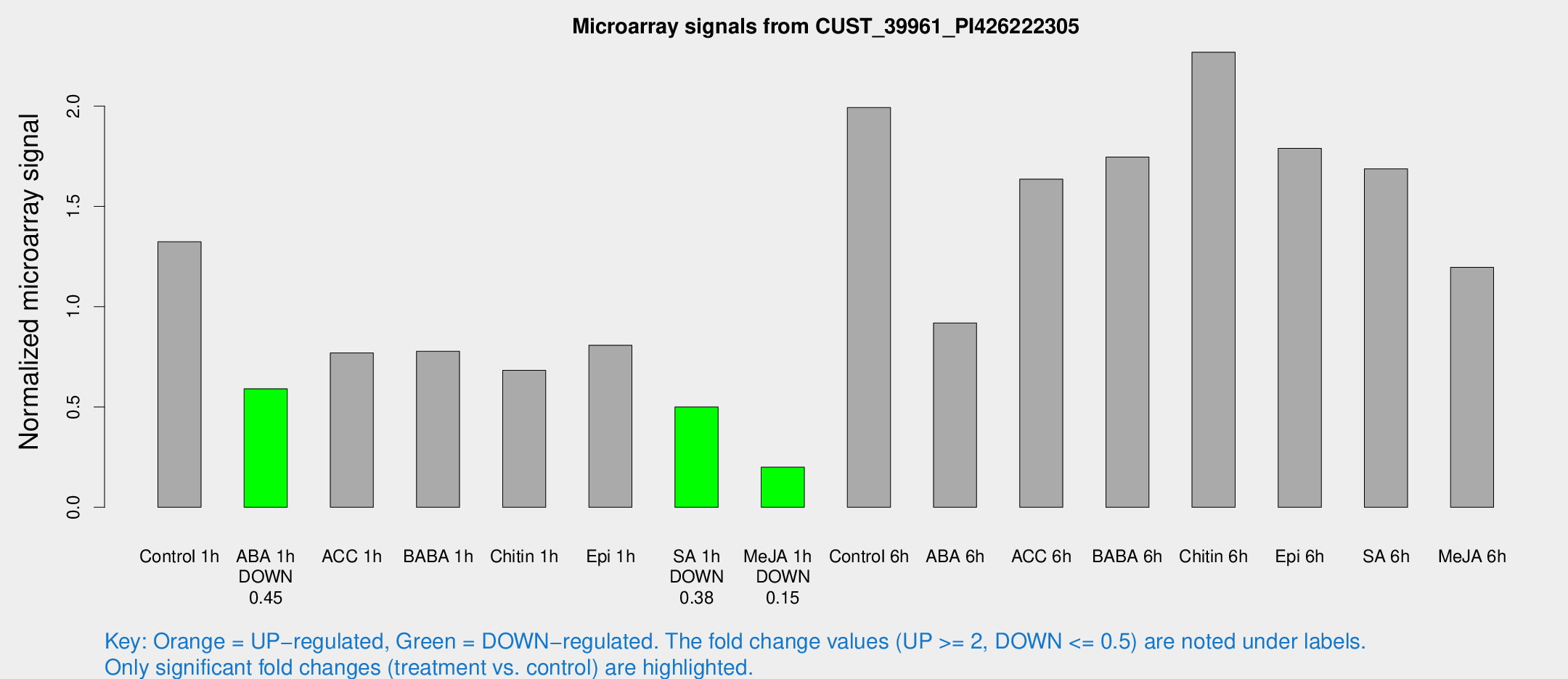
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 370.915 | 53.8044 | 1.32387 | 0.106819 |
| ABA 1h | 146.275 | 21.4946 | 0.590923 | 0.091286 |
| ACC 1h | 223.283 | 37.6803 | 0.769844 | 0.0788008 |
| BABA 1h | 223.419 | 56.3191 | 0.777372 | 0.161489 |
| Chitin 1h | 168.403 | 10.3431 | 0.682877 | 0.0578464 |
| Epi 1h | 194.894 | 23.6826 | 0.80775 | 0.0949238 |
| SA 1h | 149.038 | 33.9557 | 0.499812 | 0.119168 |
| Me-JA 1h | 45.9276 | 7.52437 | 0.200092 | 0.0197293 |
| Control 6h | 575.206 | 125.718 | 1.99253 | 0.322481 |
| ABA 6h | 267.99 | 15.9366 | 0.918887 | 0.0646884 |
| ACC 6h | 525.133 | 70.6218 | 1.63593 | 0.173471 |
| BABA 6h | 549.079 | 88.3808 | 1.74613 | 0.269567 |
| Chitin 6h | 668.441 | 64.8433 | 2.26857 | 0.145969 |
| Epi 6h | 562.068 | 67.8399 | 1.78952 | 0.148556 |
| SA 6h | 482.26 | 99.6281 | 1.68695 | 0.204594 |
| Me-JA 6h | 358.98 | 97.8718 | 1.19664 | 0.300409 |
Source Transcript PGSC0003DMT400081473 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G12900.1 | +3 | 2e-149 | 449 | 240/396 (61%) | unknown protein; FUNCTIONS IN: molecular_function unknown; INVOLVED IN: biological_process unknown; LOCATED IN: chloroplast; EXPRESSED IN: 21 plant structures; EXPRESSED DURING: 13 growth stages; BEST Arabidopsis thaliana protein match is: unknown protein (TAIR:AT1G12330.1); Has 1807 Blast hits to 1807 proteins in 277 species: Archae - 0; Bacteria - 0; Metazoa - 736; Fungi - 347; Plants - 385; Viruses - 0; Other Eukaryotes - 339 (source: NCBI BLink). | chr5:4072151-4074445 REVERSE LENGTH=562 |
