Probe CUST_39481_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_39481_PI426222305 | JHI_St_60k_v1 | DMT400083907 | GGGTGATAATCGAATTCTTGATAAGACCATTGAATTTGACCAAGTTGACAGAGAAAGAAA |
All Microarray Probes Designed to Gene DMG400033650
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_39479_PI426222305 | JHI_St_60k_v1 | DMT400083906 | ATTCAATTCCCAAAGGAACTACCCAAGTGGTCCTTGGAGGCCAGTTGTCAAAAGTTGCTA |
| CUST_39518_PI426222305 | JHI_St_60k_v1 | DMT400083908 | CAAGTTGACAGAGAAAGAAATTGCTCATAGCGAAAGGCATAGAGTTGTTGGTTTAGCAAA |
| CUST_39481_PI426222305 | JHI_St_60k_v1 | DMT400083907 | GGGTGATAATCGAATTCTTGATAAGACCATTGAATTTGACCAAGTTGACAGAGAAAGAAA |
Microarray Signals from CUST_39481_PI426222305
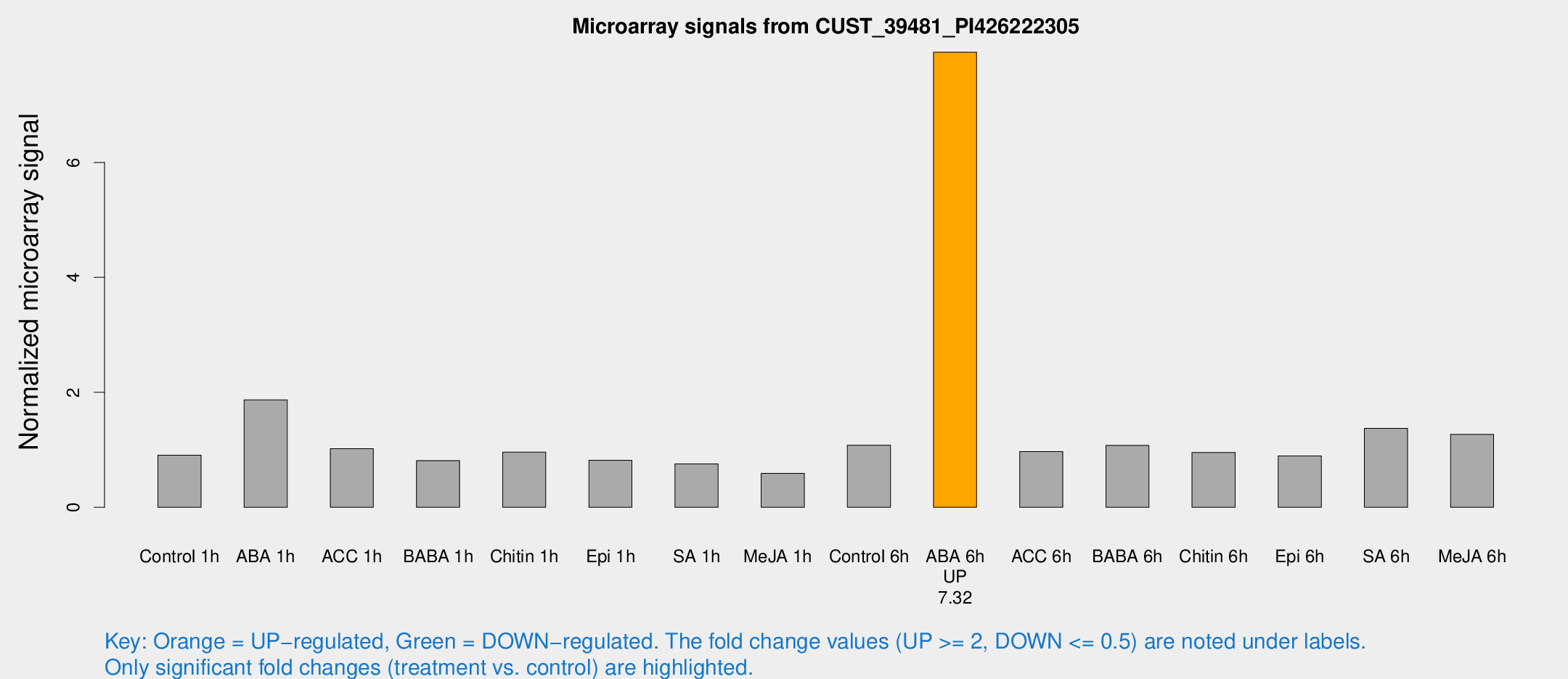
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 133.866 | 25.5054 | 0.90766 | 0.113978 |
| ABA 1h | 237.365 | 22.2519 | 1.87027 | 0.168122 |
| ACC 1h | 159.828 | 39.9326 | 1.02033 | 0.205598 |
| BABA 1h | 115.502 | 23.0211 | 0.81219 | 0.1191 |
| Chitin 1h | 123.882 | 11.9734 | 0.960115 | 0.129594 |
| Epi 1h | 102.674 | 14.9274 | 0.817591 | 0.0958086 |
| SA 1h | 111.981 | 13.9515 | 0.754336 | 0.0502482 |
| Me-JA 1h | 68.9309 | 5.21195 | 0.591907 | 0.0677502 |
| Control 6h | 181.688 | 61.4153 | 1.08121 | 0.418203 |
| ABA 6h | 1248.29 | 240.578 | 7.91687 | 1.279 |
| ACC 6h | 166.109 | 38.0251 | 0.970545 | 0.0851695 |
| BABA 6h | 177.203 | 33.994 | 1.07614 | 0.194846 |
| Chitin 6h | 145.577 | 13.0894 | 0.95374 | 0.118479 |
| Epi 6h | 151.692 | 32.3214 | 0.894443 | 0.299489 |
| SA 6h | 200.083 | 35.148 | 1.37391 | 0.425677 |
| Me-JA 6h | 200.013 | 64.2842 | 1.26882 | 0.326443 |
Source Transcript PGSC0003DMT400083907 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G02205.3 | +1 | 0.0 | 644 | 322/480 (67%) | Fatty acid hydroxylase superfamily | chr1:418818-422154 FORWARD LENGTH=630 |
