Probe CUST_39479_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_39479_PI426222305 | JHI_St_60k_v1 | DMT400083906 | ATTCAATTCCCAAAGGAACTACCCAAGTGGTCCTTGGAGGCCAGTTGTCAAAAGTTGCTA |
All Microarray Probes Designed to Gene DMG400033650
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_39479_PI426222305 | JHI_St_60k_v1 | DMT400083906 | ATTCAATTCCCAAAGGAACTACCCAAGTGGTCCTTGGAGGCCAGTTGTCAAAAGTTGCTA |
| CUST_39518_PI426222305 | JHI_St_60k_v1 | DMT400083908 | CAAGTTGACAGAGAAAGAAATTGCTCATAGCGAAAGGCATAGAGTTGTTGGTTTAGCAAA |
| CUST_39481_PI426222305 | JHI_St_60k_v1 | DMT400083907 | GGGTGATAATCGAATTCTTGATAAGACCATTGAATTTGACCAAGTTGACAGAGAAAGAAA |
Microarray Signals from CUST_39479_PI426222305
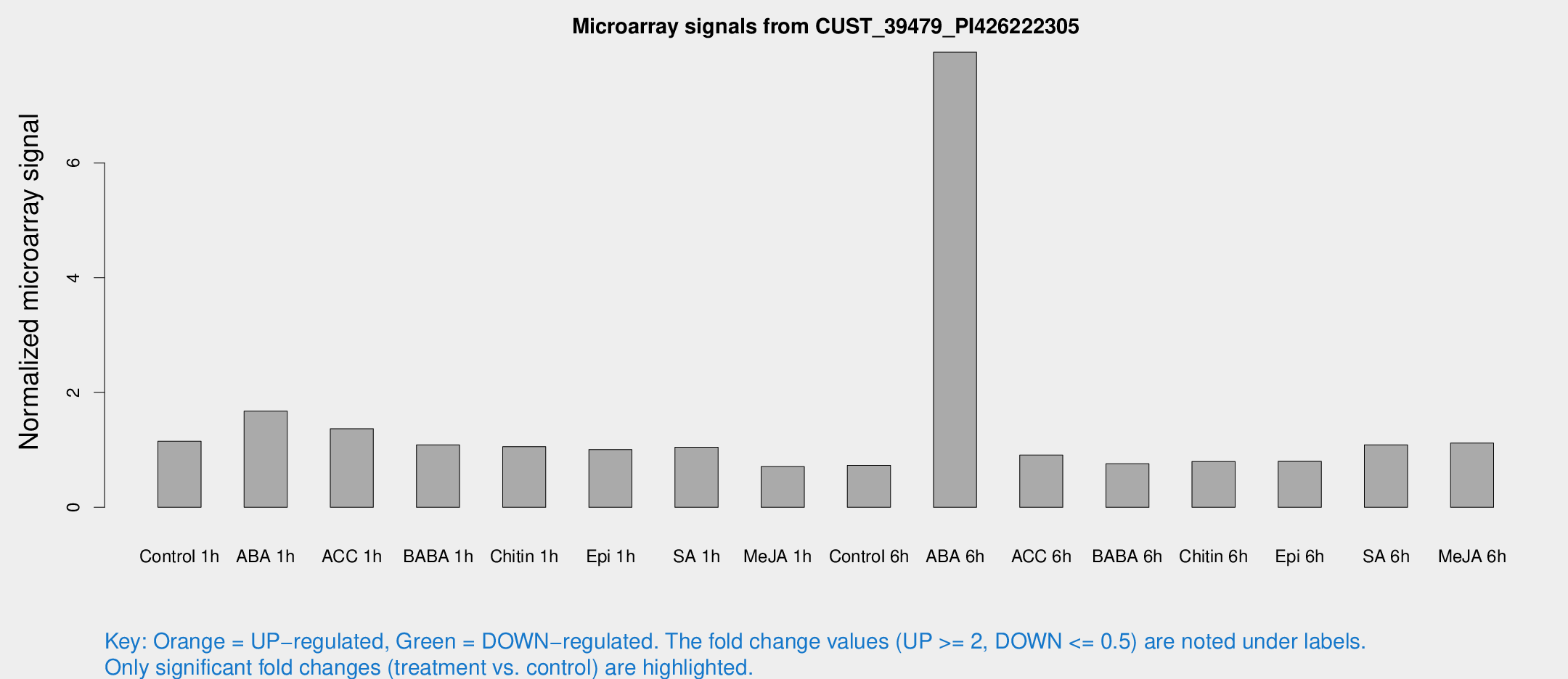
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 1611.46 | 302.314 | 1.1505 | 0.134294 |
| ABA 1h | 2022.9 | 180.747 | 1.67433 | 0.0967 |
| ACC 1h | 2097.04 | 651.677 | 1.36916 | 0.346622 |
| BABA 1h | 1507.82 | 350.546 | 1.08755 | 0.172389 |
| Chitin 1h | 1313.67 | 183.542 | 1.05661 | 0.124378 |
| Epi 1h | 1179.82 | 68.1912 | 1.00605 | 0.0581417 |
| SA 1h | 1505.86 | 251.191 | 1.04752 | 0.140962 |
| Me-JA 1h | 791.344 | 72.3812 | 0.709792 | 0.0943089 |
| Control 6h | 1273.65 | 500.759 | 0.730551 | 0.407756 |
| ABA 6h | 12172.3 | 2744.96 | 7.92935 | 1.85042 |
| ACC 6h | 1469.71 | 281.074 | 0.909782 | 0.127788 |
| BABA 6h | 1209.95 | 284.84 | 0.758246 | 0.163928 |
| Chitin 6h | 1158.74 | 113.669 | 0.795955 | 0.090707 |
| Epi 6h | 1300.61 | 306.735 | 0.801606 | 0.253701 |
| SA 6h | 1456.1 | 84.141 | 1.08647 | 0.124682 |
| Me-JA 6h | 1628.8 | 411.229 | 1.11862 | 0.235385 |
Source Transcript PGSC0003DMT400083906 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G02190.1 | +1 | 2e-78 | 253 | 113/209 (54%) | Fatty acid hydroxylase superfamily | chr1:415154-417796 FORWARD LENGTH=627 |
