Probe CUST_39337_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_39337_PI426222305 | JHI_St_60k_v1 | DMT400012831 | ACTTTCTGCGAAAAGACTTTGCTAACAATGGAACAGACTAATTGGTATTCCTGGTGCACT |
All Microarray Probes Designed to Gene DMG400004996
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_39274_PI426222305 | JHI_St_60k_v1 | DMT400012828 | CCAGTTCAGTGAGTTTAACTTTTTATAAATGGAAAGAGAGACATATGACACATTCGAGCA |
| CUST_39291_PI426222305 | JHI_St_60k_v1 | DMT400012826 | TACAATGTGGGTTCTTTTTCTCTTTTGAATCGGTGATTTTATTGAATATGTGGAGTCGGG |
| CUST_39310_PI426222305 | JHI_St_60k_v1 | DMT400012829 | GGCTTTTTATGTCTTTTATTTAGTAGCTGGAAAGTGTATGAACATGTCTACAACTCACTG |
| CUST_39337_PI426222305 | JHI_St_60k_v1 | DMT400012831 | ACTTTCTGCGAAAAGACTTTGCTAACAATGGAACAGACTAATTGGTATTCCTGGTGCACT |
Microarray Signals from CUST_39337_PI426222305
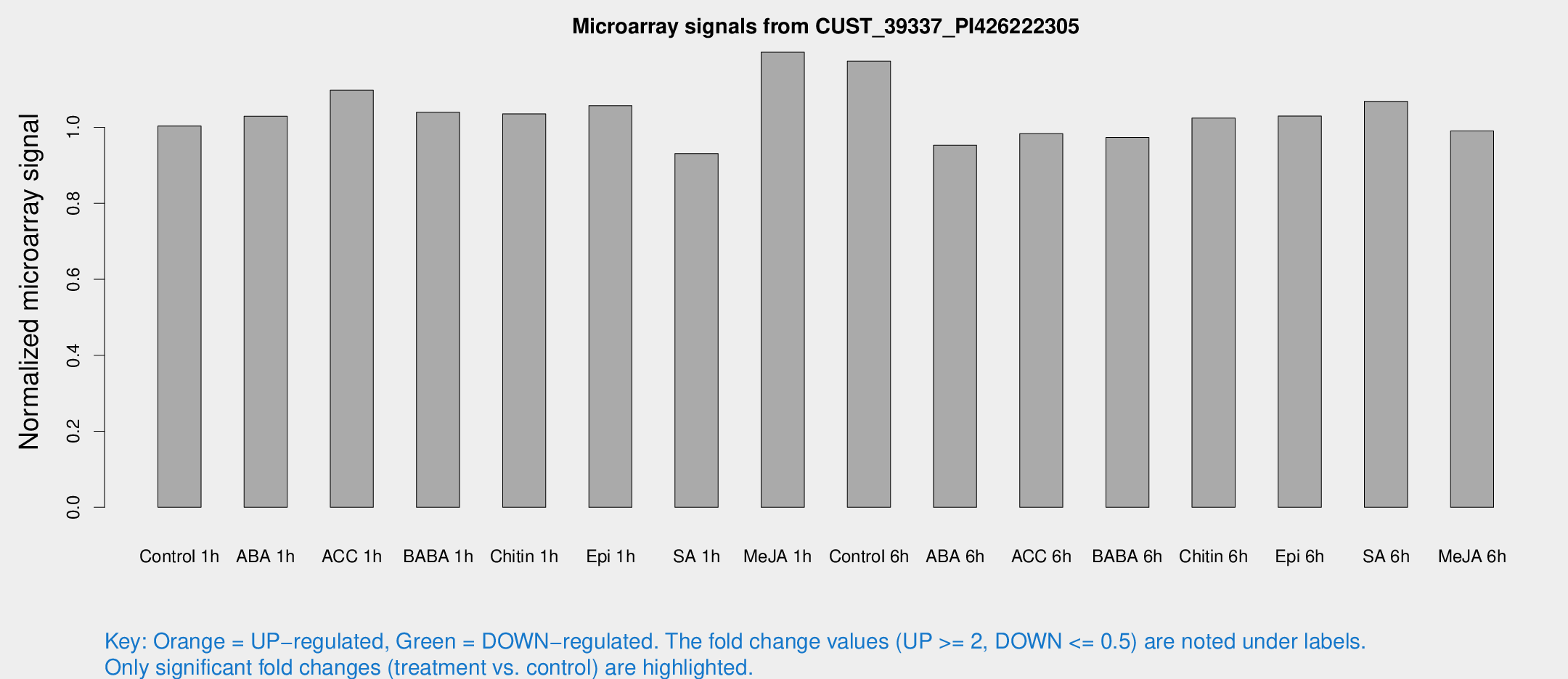
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 6.49586 | 3.53848 | 1.00373 | 0.550349 |
| ABA 1h | 5.86772 | 3.40335 | 1.02947 | 0.596687 |
| ACC 1h | 7.46197 | 4.45223 | 1.09785 | 0.636565 |
| BABA 1h | 6.46286 | 3.75311 | 1.03996 | 0.603122 |
| Chitin 1h | 6.02175 | 3.50308 | 1.03562 | 0.599838 |
| Epi 1h | 5.90863 | 3.43122 | 1.05725 | 0.612222 |
| SA 1h | 6.1632 | 3.567 | 0.931165 | 0.538577 |
| Me-JA 1h | 6.31663 | 3.67756 | 1.19785 | 0.694936 |
| Control 6h | 7.90476 | 3.69905 | 1.17461 | 0.591635 |
| ABA 6h | 6.52622 | 3.78923 | 0.952964 | 0.553064 |
| ACC 6h | 7.43268 | 4.40925 | 0.983556 | 0.56956 |
| BABA 6h | 7.03438 | 4.08502 | 0.973554 | 0.563867 |
| Chitin 6h | 7.0214 | 4.06601 | 1.02439 | 0.593146 |
| Epi 6h | 7.57866 | 4.45551 | 1.02984 | 0.597093 |
| SA 6h | 6.81007 | 3.95353 | 1.06872 | 0.620092 |
| Me-JA 6h | 6.35327 | 3.67995 | 0.990692 | 0.573687 |
Source Transcript PGSC0003DMT400012831 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G10960.1 | +1 | 9e-27 | 108 | 45/59 (76%) | UDP-D-glucose/UDP-D-galactose 4-epimerase 5 | chr4:6716083-6718472 REVERSE LENGTH=351 |
