Probe CUST_39310_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_39310_PI426222305 | JHI_St_60k_v1 | DMT400012829 | GGCTTTTTATGTCTTTTATTTAGTAGCTGGAAAGTGTATGAACATGTCTACAACTCACTG |
All Microarray Probes Designed to Gene DMG400004996
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_39274_PI426222305 | JHI_St_60k_v1 | DMT400012828 | CCAGTTCAGTGAGTTTAACTTTTTATAAATGGAAAGAGAGACATATGACACATTCGAGCA |
| CUST_39291_PI426222305 | JHI_St_60k_v1 | DMT400012826 | TACAATGTGGGTTCTTTTTCTCTTTTGAATCGGTGATTTTATTGAATATGTGGAGTCGGG |
| CUST_39310_PI426222305 | JHI_St_60k_v1 | DMT400012829 | GGCTTTTTATGTCTTTTATTTAGTAGCTGGAAAGTGTATGAACATGTCTACAACTCACTG |
| CUST_39337_PI426222305 | JHI_St_60k_v1 | DMT400012831 | ACTTTCTGCGAAAAGACTTTGCTAACAATGGAACAGACTAATTGGTATTCCTGGTGCACT |
Microarray Signals from CUST_39310_PI426222305
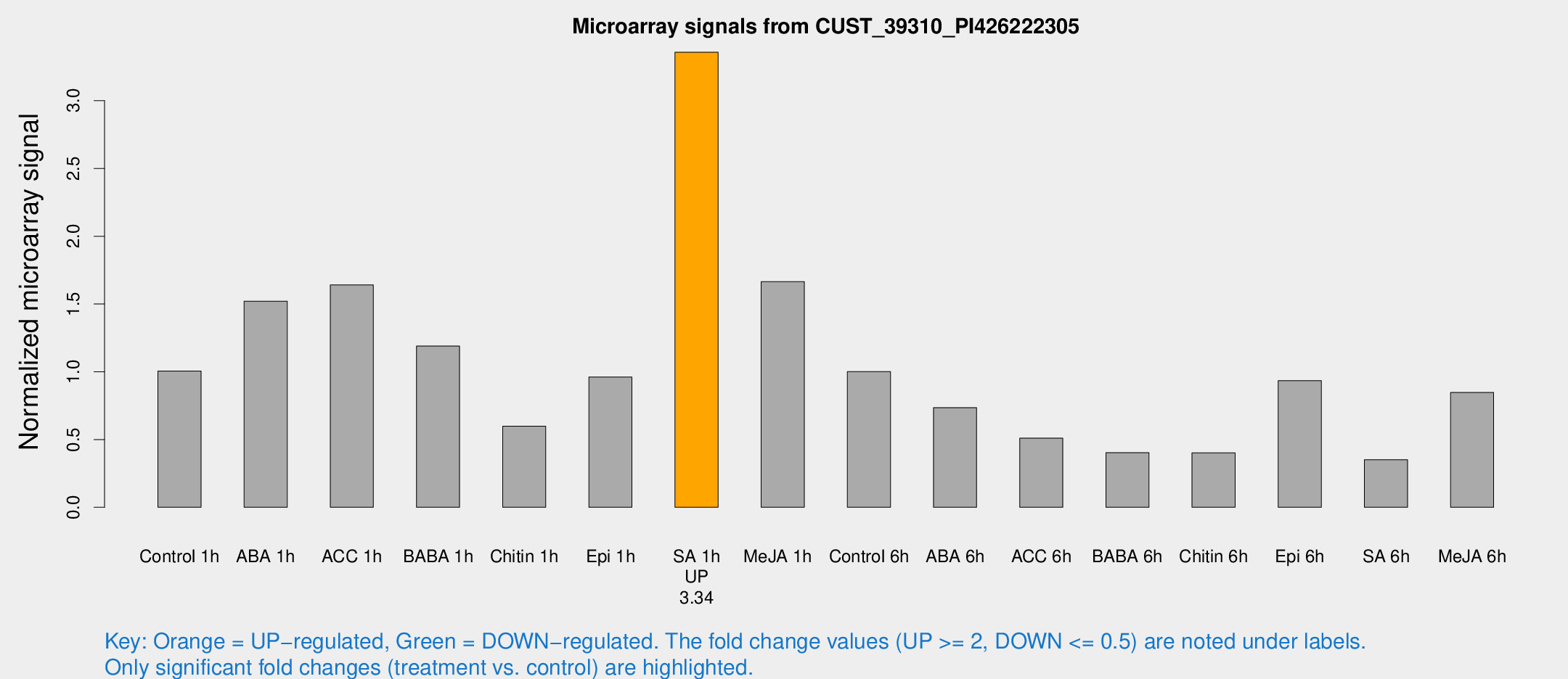
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 19.2264 | 3.33876 | 1.0052 | 0.177769 |
| ABA 1h | 25.6824 | 3.37503 | 1.52111 | 0.202213 |
| ACC 1h | 32.0895 | 4.11605 | 1.64102 | 0.216279 |
| BABA 1h | 22.2437 | 3.54471 | 1.18983 | 0.31267 |
| Chitin 1h | 11.8025 | 4.60527 | 0.598601 | 0.27432 |
| Epi 1h | 15.7999 | 3.22392 | 0.961405 | 0.197272 |
| SA 1h | 68.6804 | 13.7106 | 3.35758 | 0.614448 |
| Me-JA 1h | 25.9263 | 3.55946 | 1.66465 | 0.230829 |
| Control 6h | 24.1814 | 9.08886 | 1.00149 | 0.535484 |
| ABA 6h | 17.8961 | 6.28557 | 0.735583 | 0.488894 |
| ACC 6h | 11.7872 | 4.06018 | 0.510381 | 0.185002 |
| BABA 6h | 9.91443 | 3.99193 | 0.403133 | 0.190453 |
| Chitin 6h | 8.2311 | 3.60808 | 0.40197 | 0.182719 |
| Epi 6h | 26.1345 | 12.3158 | 0.934597 | 0.718477 |
| SA 6h | 6.73344 | 3.40184 | 0.351563 | 0.183809 |
| Me-JA 6h | 16.1588 | 3.36352 | 0.847411 | 0.178785 |
Source Transcript PGSC0003DMT400012829 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G23920.1 | +3 | 0.0 | 579 | 273/344 (79%) | UDP-D-glucose/UDP-D-galactose 4-epimerase 2 | chr4:12431416-12433666 FORWARD LENGTH=350 |
