Probe CUST_38898_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_38898_PI426222305 | JHI_St_60k_v1 | DMT400037859 | CTGAAACTTTGAGTAAGCTTTGGATTGACAATGGAAGGAGCTACAAAATGATATTTGCAG |
All Microarray Probes Designed to Gene DMG400014604
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_38897_PI426222305 | JHI_St_60k_v1 | DMT400037856 | TTTGATCTTGTTGAGGAAATACGTCCATGCAGAGACTTAAAGAGGCTAGTGCGGAGACCA |
| CUST_38918_PI426222305 | JHI_St_60k_v1 | DMT400037858 | CTGAAACTTTGAGTAAGCTTTGGATTGACAATGGAAGGAGCTACAAAATGATATTTGCAG |
| CUST_38898_PI426222305 | JHI_St_60k_v1 | DMT400037859 | CTGAAACTTTGAGTAAGCTTTGGATTGACAATGGAAGGAGCTACAAAATGATATTTGCAG |
| CUST_38923_PI426222305 | JHI_St_60k_v1 | DMT400037857 | AACGGTGAAGCTTGAGTACTCCTGGTGTGTAATTTTGTTCTTGACAGATGTGGGAGGTAC |
Microarray Signals from CUST_38898_PI426222305
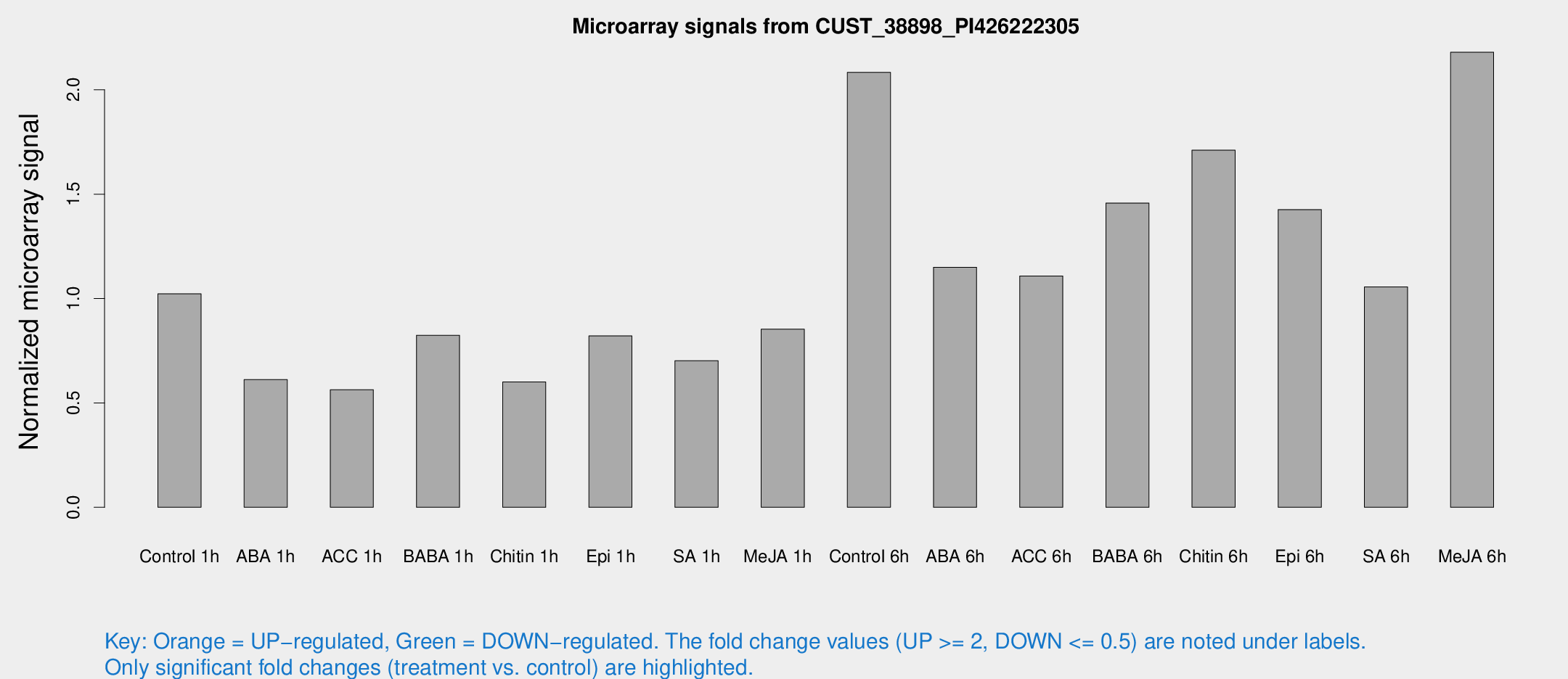
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 61.204 | 4.76341 | 1.02252 | 0.0797451 |
| ABA 1h | 34.8441 | 8.40316 | 0.611866 | 0.146407 |
| ACC 1h | 37.7329 | 10.3141 | 0.563379 | 0.140053 |
| BABA 1h | 49.2017 | 9.19522 | 0.824225 | 0.0900373 |
| Chitin 1h | 35.635 | 10.8994 | 0.600565 | 0.15082 |
| Epi 1h | 43.1282 | 5.18007 | 0.820858 | 0.0921054 |
| SA 1h | 45.7304 | 10.0384 | 0.702259 | 0.143163 |
| Me-JA 1h | 41.7139 | 4.12586 | 0.853515 | 0.0842214 |
| Control 6h | 134.334 | 34.271 | 2.0835 | 0.433021 |
| ABA 6h | 75.4228 | 12.6132 | 1.14991 | 0.158332 |
| ACC 6h | 77.7812 | 12.1977 | 1.10823 | 0.0863988 |
| BABA 6h | 101.589 | 19.8399 | 1.45742 | 0.265759 |
| Chitin 6h | 111.322 | 17.5349 | 1.71047 | 0.307431 |
| Epi 6h | 108.767 | 37.272 | 1.42622 | 0.632221 |
| SA 6h | 63.171 | 6.76132 | 1.05569 | 0.0879379 |
| Me-JA 6h | 140.591 | 40.3318 | 2.17982 | 0.449312 |
Source Transcript PGSC0003DMT400037859 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G37710.1 | +3 | 0.0 | 564 | 293/451 (65%) | alpha/beta-Hydrolases superfamily protein | chr5:14979159-14981400 FORWARD LENGTH=436 |
