Probe CUST_38923_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_38923_PI426222305 | JHI_St_60k_v1 | DMT400037857 | AACGGTGAAGCTTGAGTACTCCTGGTGTGTAATTTTGTTCTTGACAGATGTGGGAGGTAC |
All Microarray Probes Designed to Gene DMG400014604
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_38897_PI426222305 | JHI_St_60k_v1 | DMT400037856 | TTTGATCTTGTTGAGGAAATACGTCCATGCAGAGACTTAAAGAGGCTAGTGCGGAGACCA |
| CUST_38918_PI426222305 | JHI_St_60k_v1 | DMT400037858 | CTGAAACTTTGAGTAAGCTTTGGATTGACAATGGAAGGAGCTACAAAATGATATTTGCAG |
| CUST_38898_PI426222305 | JHI_St_60k_v1 | DMT400037859 | CTGAAACTTTGAGTAAGCTTTGGATTGACAATGGAAGGAGCTACAAAATGATATTTGCAG |
| CUST_38923_PI426222305 | JHI_St_60k_v1 | DMT400037857 | AACGGTGAAGCTTGAGTACTCCTGGTGTGTAATTTTGTTCTTGACAGATGTGGGAGGTAC |
Microarray Signals from CUST_38923_PI426222305
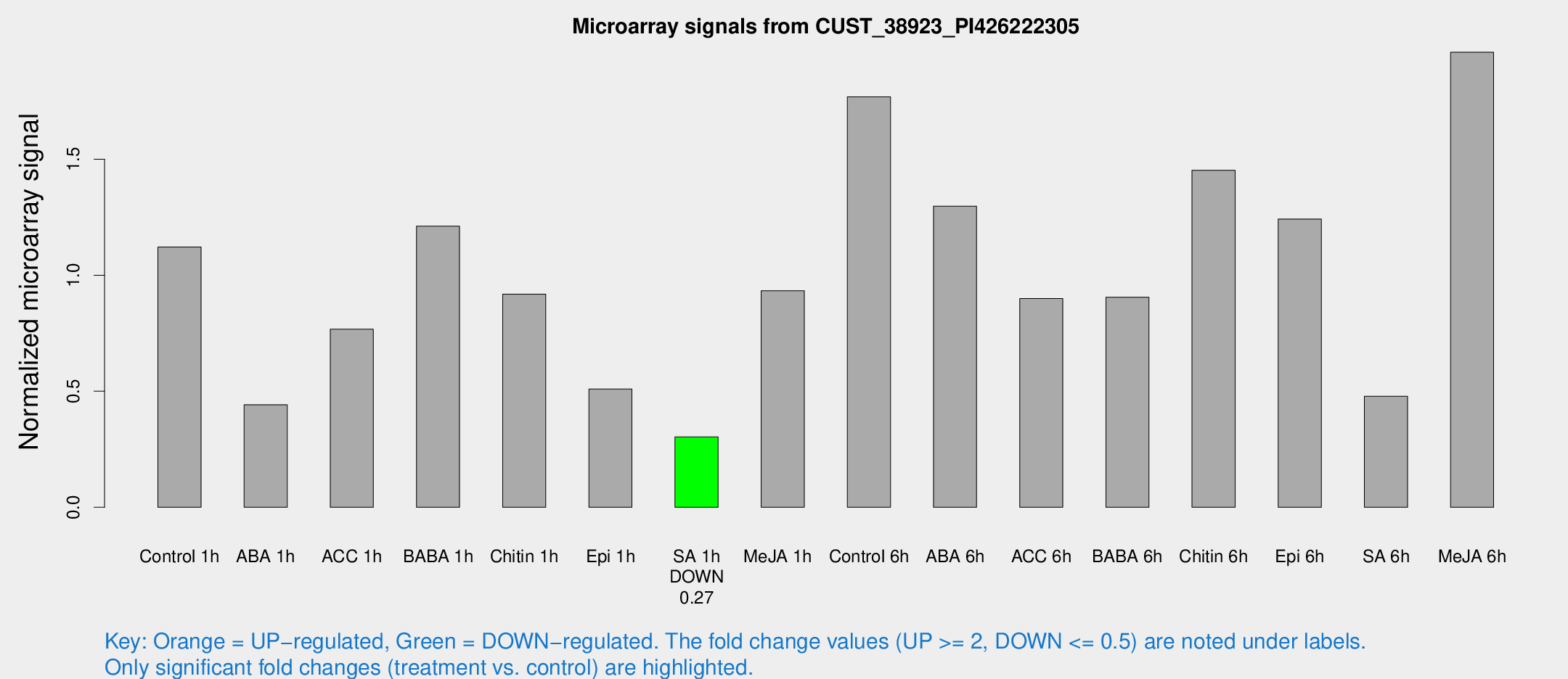
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 29.3522 | 3.43282 | 1.12136 | 0.132478 |
| ABA 1h | 11.2302 | 3.02614 | 0.44191 | 0.194565 |
| ACC 1h | 22.1553 | 6.26062 | 0.767261 | 0.200844 |
| BABA 1h | 31.0737 | 4.4946 | 1.21108 | 0.186953 |
| Chitin 1h | 22.7576 | 4.83477 | 0.918698 | 0.316992 |
| Epi 1h | 14.0419 | 6.15754 | 0.509504 | 0.266783 |
| SA 1h | 8.28551 | 3.05793 | 0.303149 | 0.116969 |
| Me-JA 1h | 19.9313 | 3.32497 | 0.932897 | 0.155023 |
| Control 6h | 46.7354 | 4.58009 | 1.76886 | 0.16203 |
| ABA 6h | 40.6774 | 12.9381 | 1.29764 | 0.474867 |
| ACC 6h | 28.3909 | 6.44115 | 0.899297 | 0.140875 |
| BABA 6h | 26.5701 | 3.91166 | 0.90513 | 0.133971 |
| Chitin 6h | 41.1807 | 5.61906 | 1.45235 | 0.173322 |
| Epi 6h | 38.0297 | 7.44948 | 1.24206 | 0.318998 |
| SA 6h | 14.0667 | 4.38073 | 0.478836 | 0.241246 |
| Me-JA 6h | 52.2156 | 8.44273 | 1.96075 | 0.170281 |
Source Transcript PGSC0003DMT400037857 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G37710.1 | +2 | 6e-34 | 132 | 75/152 (49%) | alpha/beta-Hydrolases superfamily protein | chr5:14979159-14981400 FORWARD LENGTH=436 |
