Probe CUST_38269_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_38269_PI426222305 | JHI_St_60k_v1 | DMT400067357 | TTGTAAAACAGATTCTTGGTAGATGAGACCCCTGTTCGTGTGCATTCGAATTTGGAGCAC |
All Microarray Probes Designed to Gene DMG400026189
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_38269_PI426222305 | JHI_St_60k_v1 | DMT400067357 | TTGTAAAACAGATTCTTGGTAGATGAGACCCCTGTTCGTGTGCATTCGAATTTGGAGCAC |
| CUST_38254_PI426222305 | JHI_St_60k_v1 | DMT400067358 | TCGATCATGTGGAATCAACGTCAAGTTGTATTCTTGGTAGATGAGACCCCTGTTCGTGTG |
Microarray Signals from CUST_38269_PI426222305
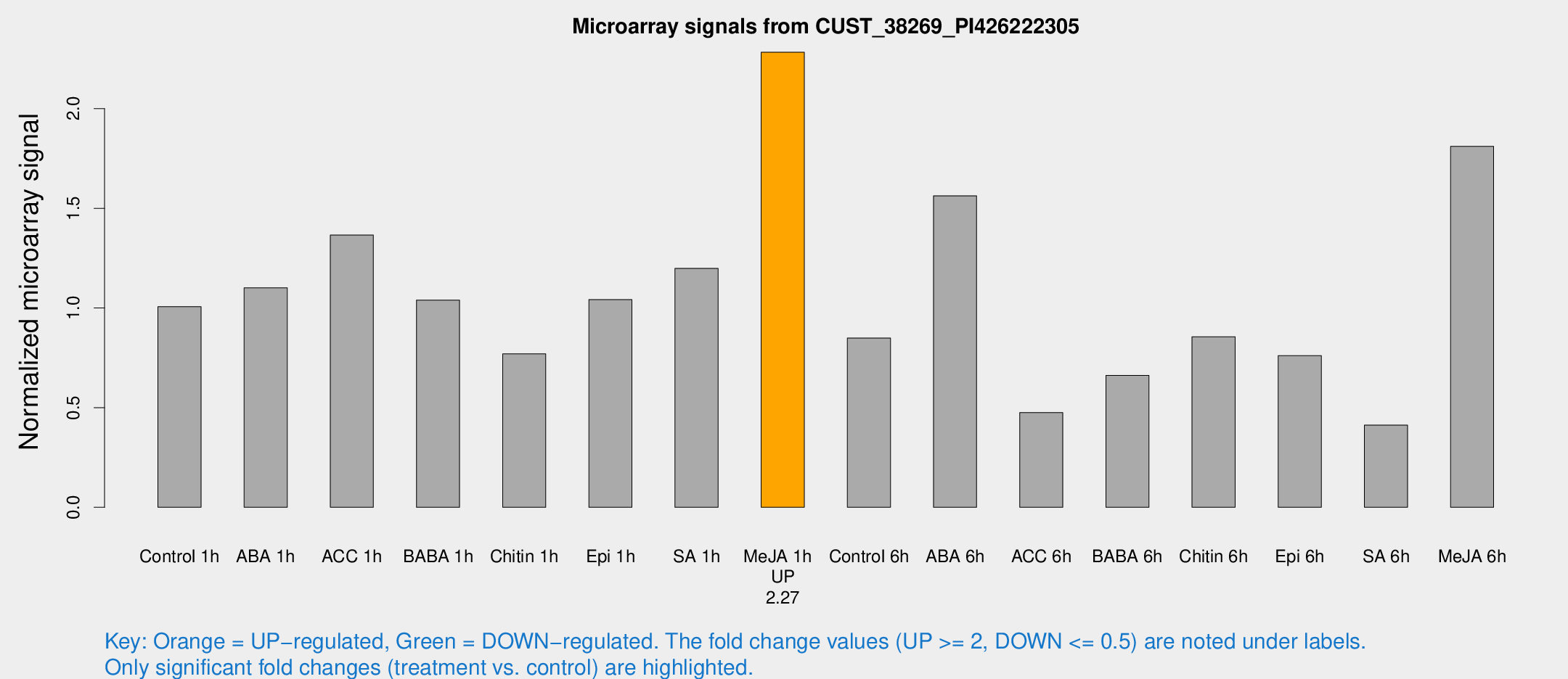
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 4964.3 | 801.469 | 1.00685 | 0.0925222 |
| ABA 1h | 4862.83 | 976.068 | 1.10114 | 0.154009 |
| ACC 1h | 7446.49 | 2005.04 | 1.36581 | 0.369583 |
| BABA 1h | 4827.14 | 279.099 | 1.03968 | 0.0738705 |
| Chitin 1h | 3377.17 | 394.397 | 0.769719 | 0.0722059 |
| Epi 1h | 4364.14 | 329.589 | 1.04202 | 0.062998 |
| SA 1h | 6570.96 | 2226.62 | 1.19864 | 0.417218 |
| Me-JA 1h | 9097.4 | 1085.1 | 2.28297 | 0.13181 |
| Control 6h | 5122.84 | 2046.56 | 0.848963 | 0.409316 |
| ABA 6h | 8237.75 | 1458.06 | 1.56277 | 0.339504 |
| ACC 6h | 2876.73 | 775.441 | 0.47532 | 0.137262 |
| BABA 6h | 3595.94 | 340.508 | 0.661451 | 0.0401196 |
| Chitin 6h | 4572.02 | 949.726 | 0.855918 | 0.195854 |
| Epi 6h | 4671.89 | 1406.95 | 0.760458 | 0.386765 |
| SA 6h | 2131.66 | 615.088 | 0.412387 | 0.0868055 |
| Me-JA 6h | 8677.83 | 501.244 | 1.81088 | 0.162768 |
Source Transcript PGSC0003DMT400067357 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G03210.1 | +3 | 2e-88 | 219 | 96/137 (70%) | xyloglucan endotransglucosylase/hydrolase 9 | chr4:1416019-1417197 FORWARD LENGTH=290 |
