Probe CUST_38254_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_38254_PI426222305 | JHI_St_60k_v1 | DMT400067358 | TCGATCATGTGGAATCAACGTCAAGTTGTATTCTTGGTAGATGAGACCCCTGTTCGTGTG |
All Microarray Probes Designed to Gene DMG400026189
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_38269_PI426222305 | JHI_St_60k_v1 | DMT400067357 | TTGTAAAACAGATTCTTGGTAGATGAGACCCCTGTTCGTGTGCATTCGAATTTGGAGCAC |
| CUST_38254_PI426222305 | JHI_St_60k_v1 | DMT400067358 | TCGATCATGTGGAATCAACGTCAAGTTGTATTCTTGGTAGATGAGACCCCTGTTCGTGTG |
Microarray Signals from CUST_38254_PI426222305
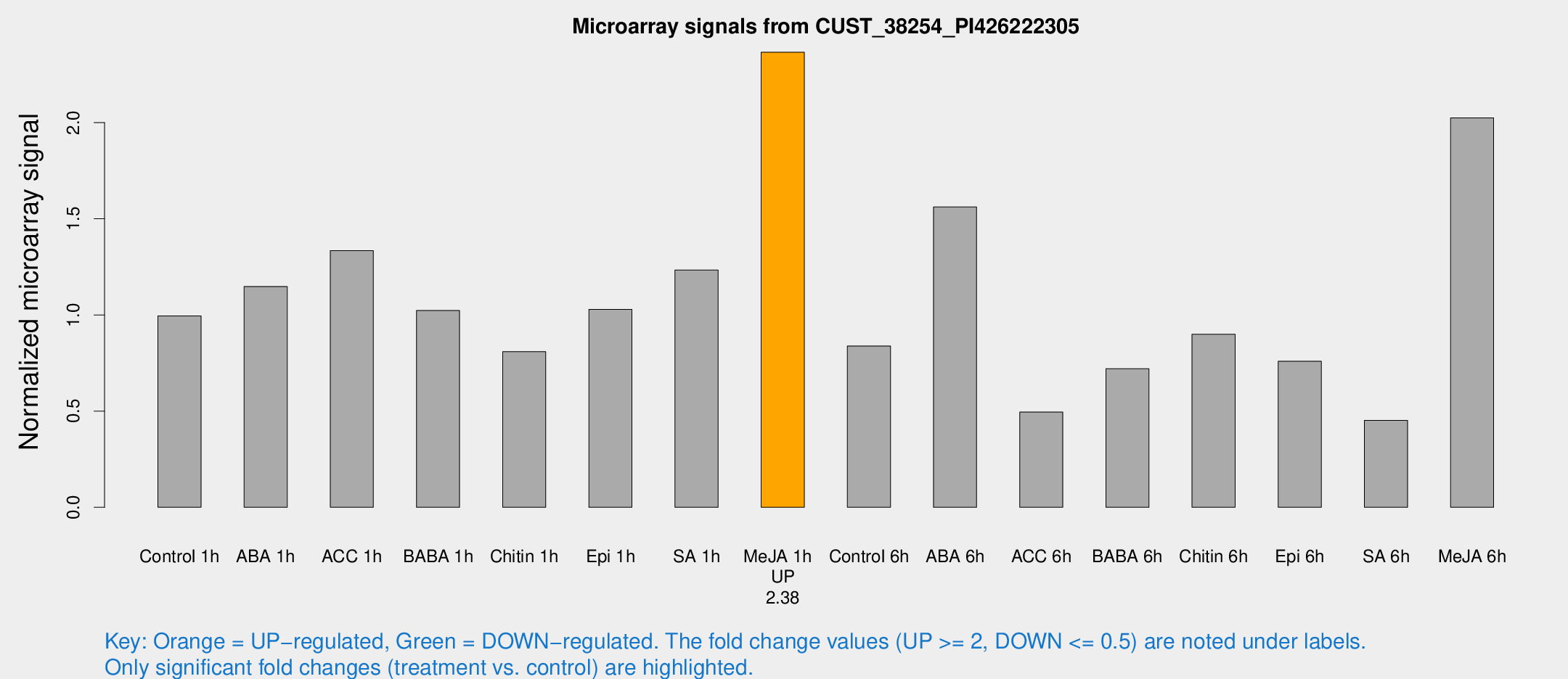
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 4702.1 | 801.68 | 0.99539 | 0.102308 |
| ABA 1h | 4870.66 | 1066.12 | 1.14806 | 0.172952 |
| ACC 1h | 7122.52 | 2100.35 | 1.33415 | 0.432943 |
| BABA 1h | 4531.47 | 261.889 | 1.0231 | 0.0784778 |
| Chitin 1h | 3370.97 | 316.866 | 0.80934 | 0.0693808 |
| Epi 1h | 4097.97 | 237.166 | 1.02894 | 0.0594114 |
| SA 1h | 6439.24 | 2153.76 | 1.23328 | 0.429912 |
| Me-JA 1h | 8998.23 | 1051.36 | 2.36596 | 0.136602 |
| Control 6h | 4864.92 | 1945.13 | 0.838754 | 0.418388 |
| ABA 6h | 7820.71 | 1263.96 | 1.56143 | 0.313607 |
| ACC 6h | 2929.6 | 895.176 | 0.495007 | 0.157485 |
| BABA 6h | 3774.97 | 498.84 | 0.720583 | 0.0784092 |
| Chitin 6h | 4567.31 | 881.554 | 0.899605 | 0.201067 |
| Epi 6h | 4493.63 | 1363.18 | 0.759625 | 0.412324 |
| SA 6h | 2243.13 | 677.159 | 0.451507 | 0.101438 |
| Me-JA 6h | 9271.88 | 536.341 | 2.02428 | 0.128073 |
Source Transcript PGSC0003DMT400067358 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G03210.1 | +3 | 5e-157 | 447 | 203/292 (70%) | xyloglucan endotransglucosylase/hydrolase 9 | chr4:1416019-1417197 FORWARD LENGTH=290 |
