Probe CUST_38026_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_38026_PI426222305 | JHI_St_60k_v1 | DMT400081397 | GTAGGCTGGCTCTTTTTGTGAAGAAATGTGTCTTGCCTACTTAAAAAGATCATAATATCA |
All Microarray Probes Designed to Gene DMG400031819
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_38041_PI426222305 | JHI_St_60k_v1 | DMT400081401 | CATTTGCAGACGGTATGTTTCAAATTGTAAATTTGGGATCATCATTCCTAGTGTCCATAT |
| CUST_38026_PI426222305 | JHI_St_60k_v1 | DMT400081397 | GTAGGCTGGCTCTTTTTGTGAAGAAATGTGTCTTGCCTACTTAAAAAGATCATAATATCA |
| CUST_38022_PI426222305 | JHI_St_60k_v1 | DMT400081398 | GTAGGCTGGCTCTTTTTGTGAAGAAATGTGTCTTGCCTACTTAAAAAGATCATAATATCA |
| CUST_38027_PI426222305 | JHI_St_60k_v1 | DMT400081399 | GTAGGCTGGCTCTTTTTGTGAAGAAATGTGTCTTGCCTACTTAAAAAGATCATAATATCA |
| CUST_38046_PI426222305 | JHI_St_60k_v1 | DMT400081396 | TCCGTAGGCTGGCTCTTTTTGTGAAGAAATGTGTCTTGCCTACTTAAAAAGATCATAATA |
Microarray Signals from CUST_38026_PI426222305
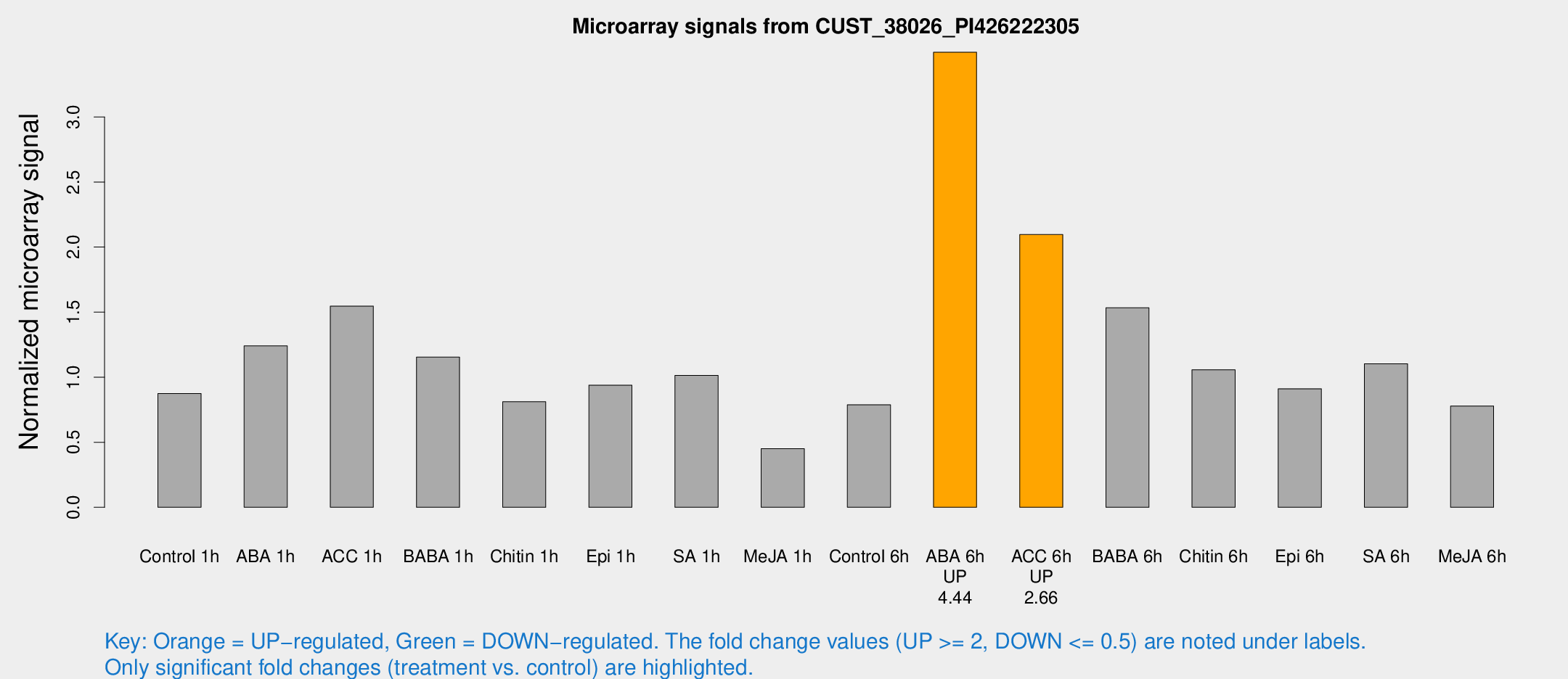
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 361.362 | 23.1868 | 0.874343 | 0.0511003 |
| ABA 1h | 513.317 | 173.264 | 1.2404 | 0.456766 |
| ACC 1h | 742.094 | 227.656 | 1.54588 | 0.510604 |
| BABA 1h | 486.291 | 111.279 | 1.15434 | 0.180794 |
| Chitin 1h | 307.885 | 49.4284 | 0.81157 | 0.0645293 |
| Epi 1h | 344.086 | 58.6966 | 0.938346 | 0.161812 |
| SA 1h | 438.209 | 67.6332 | 1.0133 | 0.0927745 |
| Me-JA 1h | 158.19 | 32.8212 | 0.451151 | 0.0540554 |
| Control 6h | 354.605 | 92.8891 | 0.787495 | 0.176499 |
| ABA 6h | 1560.95 | 210.061 | 3.49755 | 0.319835 |
| ACC 6h | 1015.85 | 169.895 | 2.09609 | 0.121301 |
| BABA 6h | 751.129 | 195.518 | 1.53298 | 0.339102 |
| Chitin 6h | 463.816 | 27.0534 | 1.05742 | 0.0616183 |
| Epi 6h | 436.376 | 77.9286 | 0.91121 | 0.141115 |
| SA 6h | 450.017 | 26.2696 | 1.10268 | 0.165058 |
| Me-JA 6h | 345.702 | 88.6154 | 0.77838 | 0.17135 |
Source Transcript PGSC0003DMT400081397 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G40940.1 | +3 | 3e-143 | 454 | 224/306 (73%) | ethylene response sensor 1 | chr2:17084635-17086819 REVERSE LENGTH=613 |
