Probe CUST_38022_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_38022_PI426222305 | JHI_St_60k_v1 | DMT400081398 | GTAGGCTGGCTCTTTTTGTGAAGAAATGTGTCTTGCCTACTTAAAAAGATCATAATATCA |
All Microarray Probes Designed to Gene DMG400031819
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_38041_PI426222305 | JHI_St_60k_v1 | DMT400081401 | CATTTGCAGACGGTATGTTTCAAATTGTAAATTTGGGATCATCATTCCTAGTGTCCATAT |
| CUST_38026_PI426222305 | JHI_St_60k_v1 | DMT400081397 | GTAGGCTGGCTCTTTTTGTGAAGAAATGTGTCTTGCCTACTTAAAAAGATCATAATATCA |
| CUST_38022_PI426222305 | JHI_St_60k_v1 | DMT400081398 | GTAGGCTGGCTCTTTTTGTGAAGAAATGTGTCTTGCCTACTTAAAAAGATCATAATATCA |
| CUST_38027_PI426222305 | JHI_St_60k_v1 | DMT400081399 | GTAGGCTGGCTCTTTTTGTGAAGAAATGTGTCTTGCCTACTTAAAAAGATCATAATATCA |
| CUST_38046_PI426222305 | JHI_St_60k_v1 | DMT400081396 | TCCGTAGGCTGGCTCTTTTTGTGAAGAAATGTGTCTTGCCTACTTAAAAAGATCATAATA |
Microarray Signals from CUST_38022_PI426222305
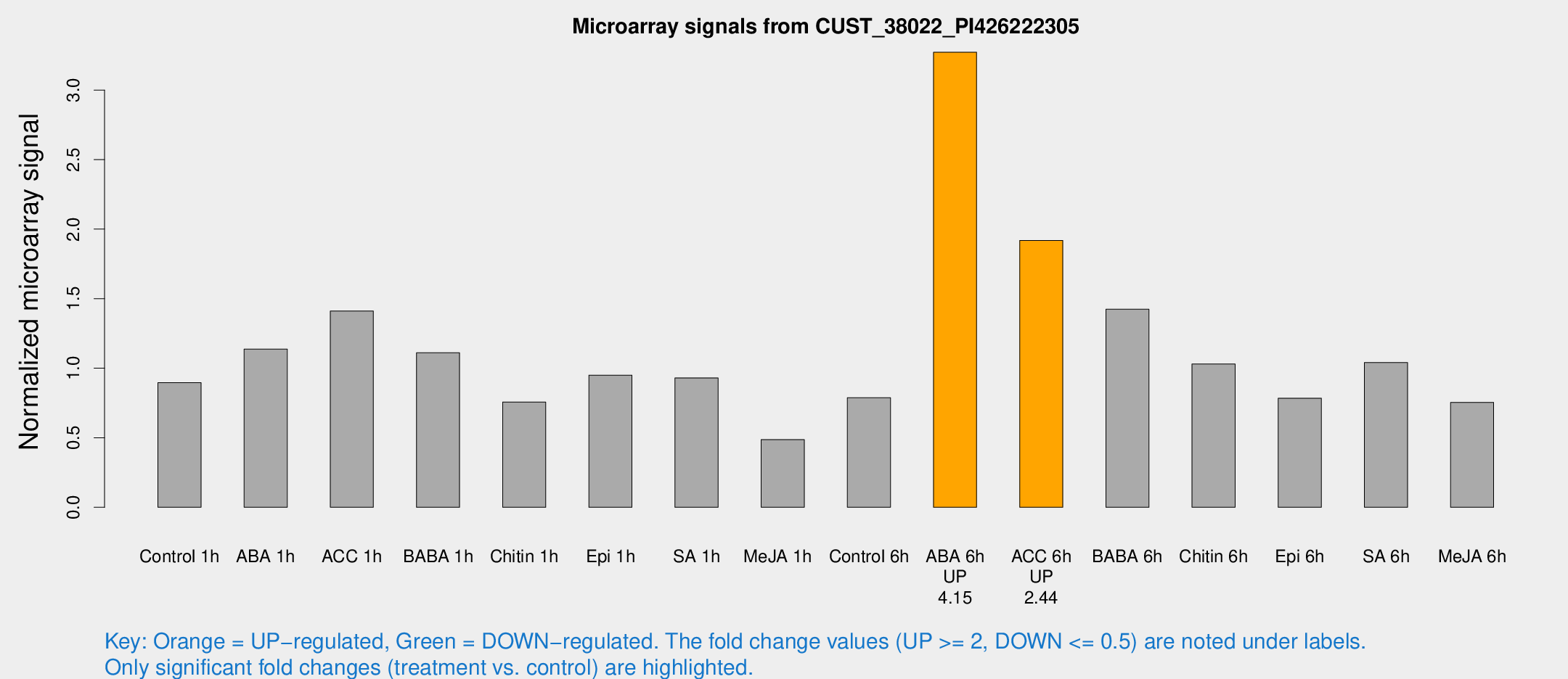
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 389.112 | 22.7754 | 0.896464 | 0.0523626 |
| ABA 1h | 480.034 | 141.71 | 1.1368 | 0.350049 |
| ACC 1h | 729.714 | 242.937 | 1.41146 | 0.530158 |
| BABA 1h | 487.372 | 105.94 | 1.11127 | 0.155194 |
| Chitin 1h | 310.243 | 71.3329 | 0.756841 | 0.112165 |
| Epi 1h | 370.519 | 70.7204 | 0.95005 | 0.191861 |
| SA 1h | 421.27 | 57.7247 | 0.930371 | 0.0756667 |
| Me-JA 1h | 176.362 | 28.0426 | 0.486425 | 0.0347415 |
| Control 6h | 371.206 | 94.6308 | 0.787857 | 0.167143 |
| ABA 6h | 1536.36 | 207.664 | 3.27189 | 0.294491 |
| ACC 6h | 974.824 | 150.731 | 1.91891 | 0.11112 |
| BABA 6h | 735.592 | 194.642 | 1.42385 | 0.31516 |
| Chitin 6h | 475.446 | 27.779 | 1.03045 | 0.0601677 |
| Epi 6h | 393.118 | 64.9118 | 0.784662 | 0.121156 |
| SA 6h | 448.47 | 32.6783 | 1.04093 | 0.184612 |
| Me-JA 6h | 355.793 | 105.03 | 0.754392 | 0.169169 |
Source Transcript PGSC0003DMT400081398 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G40940.1 | +3 | 0.0 | 713 | 377/516 (73%) | ethylene response sensor 1 | chr2:17084635-17086819 REVERSE LENGTH=613 |
