Probe CUST_36269_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_36269_PI426222305 | JHI_St_60k_v1 | DMT400039624 | GATTGGTACTTCCTTCCGTCCAAACTTTATGATACTTATTGGATTTCGAGATTCAAATAG |
All Microarray Probes Designed to Gene DMG400015327
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_36203_PI426222305 | JHI_St_60k_v1 | DMT400039625 | CACAAGGGATTGGAAAGCAACATCATTCTCACTCTTGATGACCAAGACAACTCATTCTTT |
| CUST_36269_PI426222305 | JHI_St_60k_v1 | DMT400039624 | GATTGGTACTTCCTTCCGTCCAAACTTTATGATACTTATTGGATTTCGAGATTCAAATAG |
| CUST_36316_PI426222305 | JHI_St_60k_v1 | DMT400039626 | AAATCCACTTTGAAGACAGCGCCTTTCATAGTGCGCTATTATTGTTTCACATTGCAACTT |
Microarray Signals from CUST_36269_PI426222305
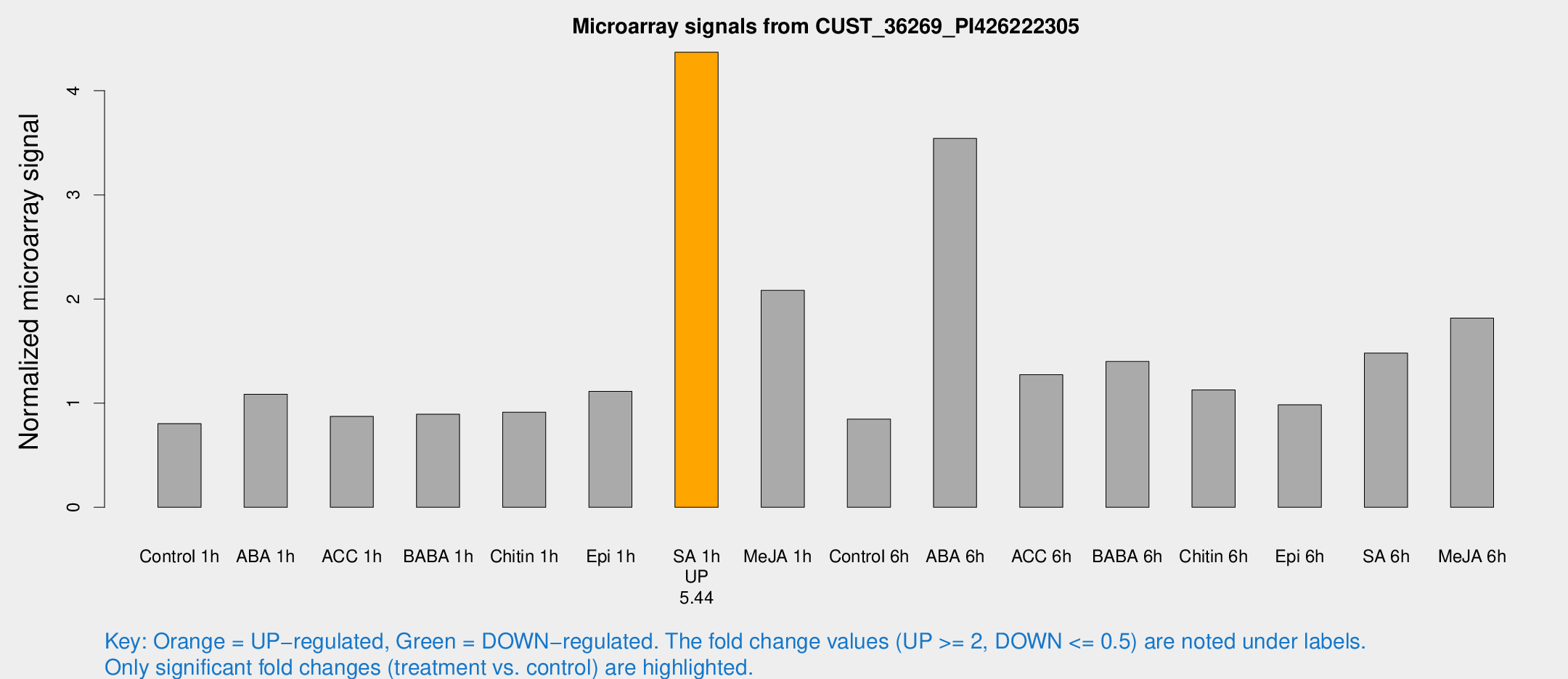
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 6.16804 | 3.57472 | 0.802846 | 0.465031 |
| ABA 1h | 7.55065 | 3.53493 | 1.08596 | 0.540387 |
| ACC 1h | 6.90468 | 4.01439 | 0.872309 | 0.505255 |
| BABA 1h | 6.6127 | 3.83316 | 0.893429 | 0.517874 |
| Chitin 1h | 6.32943 | 3.67717 | 0.913765 | 0.529223 |
| Epi 1h | 7.60119 | 3.61023 | 1.11275 | 0.560115 |
| SA 1h | 35.9531 | 7.34984 | 4.36995 | 0.636466 |
| Me-JA 1h | 13.5678 | 3.80551 | 2.08274 | 0.643663 |
| Control 6h | 6.52066 | 3.78435 | 0.847248 | 0.491674 |
| ABA 6h | 43.478 | 18.6275 | 3.54191 | 3.93672 |
| ACC 6h | 12.4459 | 4.67635 | 1.27279 | 0.589324 |
| BABA 6h | 13.4516 | 4.56856 | 1.39996 | 0.552823 |
| Chitin 6h | 9.8319 | 4.12694 | 1.12783 | 0.543821 |
| Epi 6h | 8.71111 | 4.38964 | 0.983836 | 0.509794 |
| SA 6h | 12.3026 | 3.97728 | 1.4812 | 0.586002 |
| Me-JA 6h | 16.7148 | 6.56084 | 1.81612 | 0.737372 |
Source Transcript PGSC0003DMT400039624 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G21750.1 | +3 | 8e-15 | 74 | 42/114 (37%) | UDP-glucosyl transferase 71B1 | chr3:7664565-7665986 FORWARD LENGTH=473 |
