Probe CUST_36203_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_36203_PI426222305 | JHI_St_60k_v1 | DMT400039625 | CACAAGGGATTGGAAAGCAACATCATTCTCACTCTTGATGACCAAGACAACTCATTCTTT |
All Microarray Probes Designed to Gene DMG400015327
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_36203_PI426222305 | JHI_St_60k_v1 | DMT400039625 | CACAAGGGATTGGAAAGCAACATCATTCTCACTCTTGATGACCAAGACAACTCATTCTTT |
| CUST_36269_PI426222305 | JHI_St_60k_v1 | DMT400039624 | GATTGGTACTTCCTTCCGTCCAAACTTTATGATACTTATTGGATTTCGAGATTCAAATAG |
| CUST_36316_PI426222305 | JHI_St_60k_v1 | DMT400039626 | AAATCCACTTTGAAGACAGCGCCTTTCATAGTGCGCTATTATTGTTTCACATTGCAACTT |
Microarray Signals from CUST_36203_PI426222305
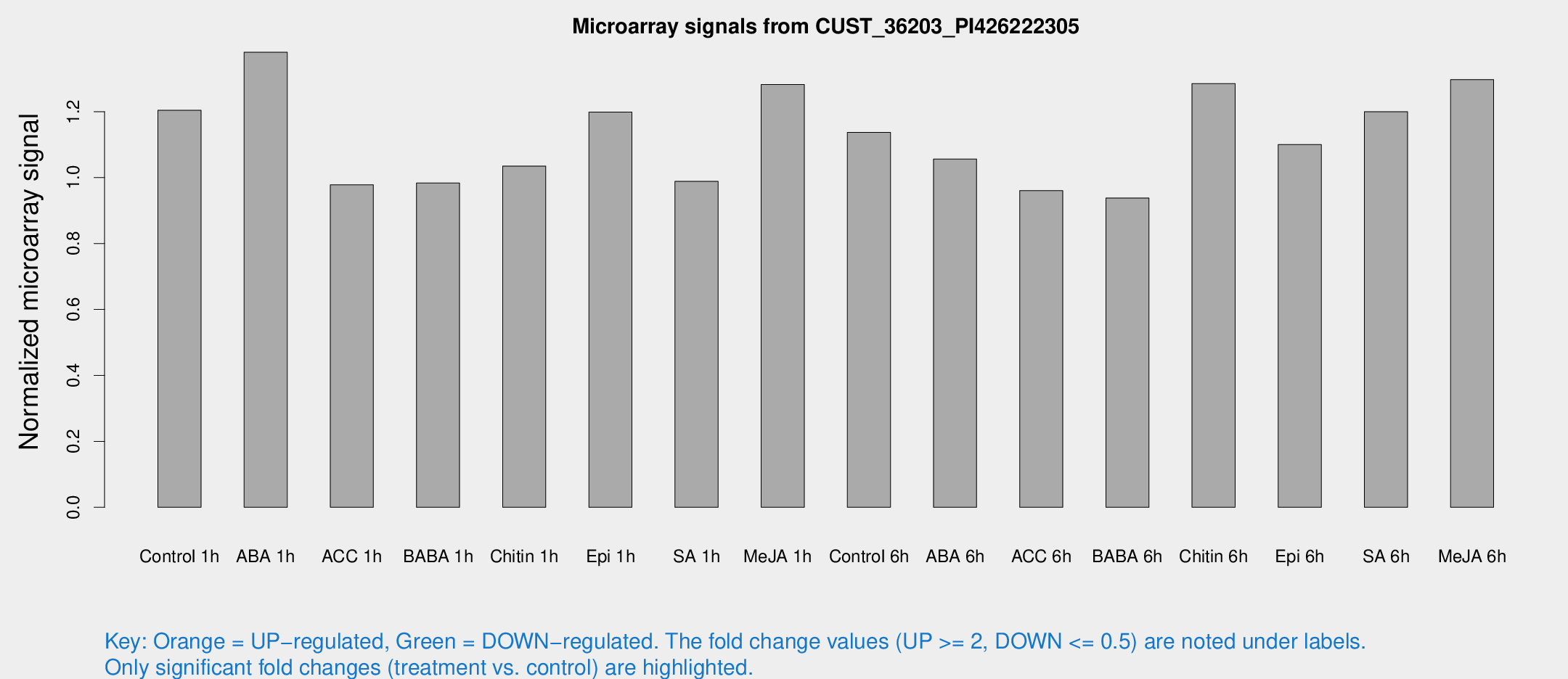
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 7.43555 | 3.01151 | 1.20412 | 0.568867 |
| ABA 1h | 7.64229 | 2.91805 | 1.38017 | 0.641706 |
| ACC 1h | 5.73117 | 3.32958 | 0.977911 | 0.567007 |
| BABA 1h | 5.40897 | 3.14025 | 0.983492 | 0.569911 |
| Chitin 1h | 5.17021 | 3.01099 | 1.03488 | 0.582983 |
| Epi 1h | 5.93251 | 2.97228 | 1.19854 | 0.604421 |
| SA 1h | 5.84697 | 2.99219 | 0.988749 | 0.51813 |
| Me-JA 1h | 5.99372 | 3.08421 | 1.28217 | 0.667796 |
| Control 6h | 6.56744 | 3.14209 | 1.13711 | 0.559217 |
| ABA 6h | 6.56552 | 3.23931 | 1.05627 | 0.54553 |
| ACC 6h | 6.40709 | 3.79709 | 0.960243 | 0.556809 |
| BABA 6h | 5.99444 | 3.48409 | 0.938013 | 0.543728 |
| Chitin 6h | 7.98289 | 3.45791 | 1.28489 | 0.584604 |
| Epi 6h | 7.49484 | 3.61442 | 1.10032 | 0.565385 |
| SA 6h | 6.90346 | 3.28572 | 1.1997 | 0.592981 |
| Me-JA 6h | 7.59986 | 3.14517 | 1.29723 | 0.57074 |
Source Transcript PGSC0003DMT400039625 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G21760.1 | +2 | 2e-140 | 422 | 236/492 (48%) | UDP-Glycosyltransferase superfamily protein | chr3:7667099-7668556 FORWARD LENGTH=485 |
