Probe CUST_33862_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_33862_PI426222305 | JHI_St_60k_v1 | DMT400012376 | TTCCTTGGTCAGACCAGTTCATCAAAAAGAGTCTCCATTCGAAACTGCTAATTGTCAATC |
All Microarray Probes Designed to Gene DMG402004850
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_33862_PI426222305 | JHI_St_60k_v1 | DMT400012376 | TTCCTTGGTCAGACCAGTTCATCAAAAAGAGTCTCCATTCGAAACTGCTAATTGTCAATC |
| CUST_33937_PI426222305 | JHI_St_60k_v1 | DMT400012377 | CAGCCAGATGAAATATAGTGTAATGTTGTTTGTTACCTGATATGCTAAAAAGTTGAGTGC |
| CUST_33837_PI426222305 | JHI_St_60k_v1 | DMT400012378 | TTTTATGTGACAGGGATTCAAATCGACCAGGCAAAGTCATTGGAGCCCTAAGCTTTGCTG |
Microarray Signals from CUST_33862_PI426222305
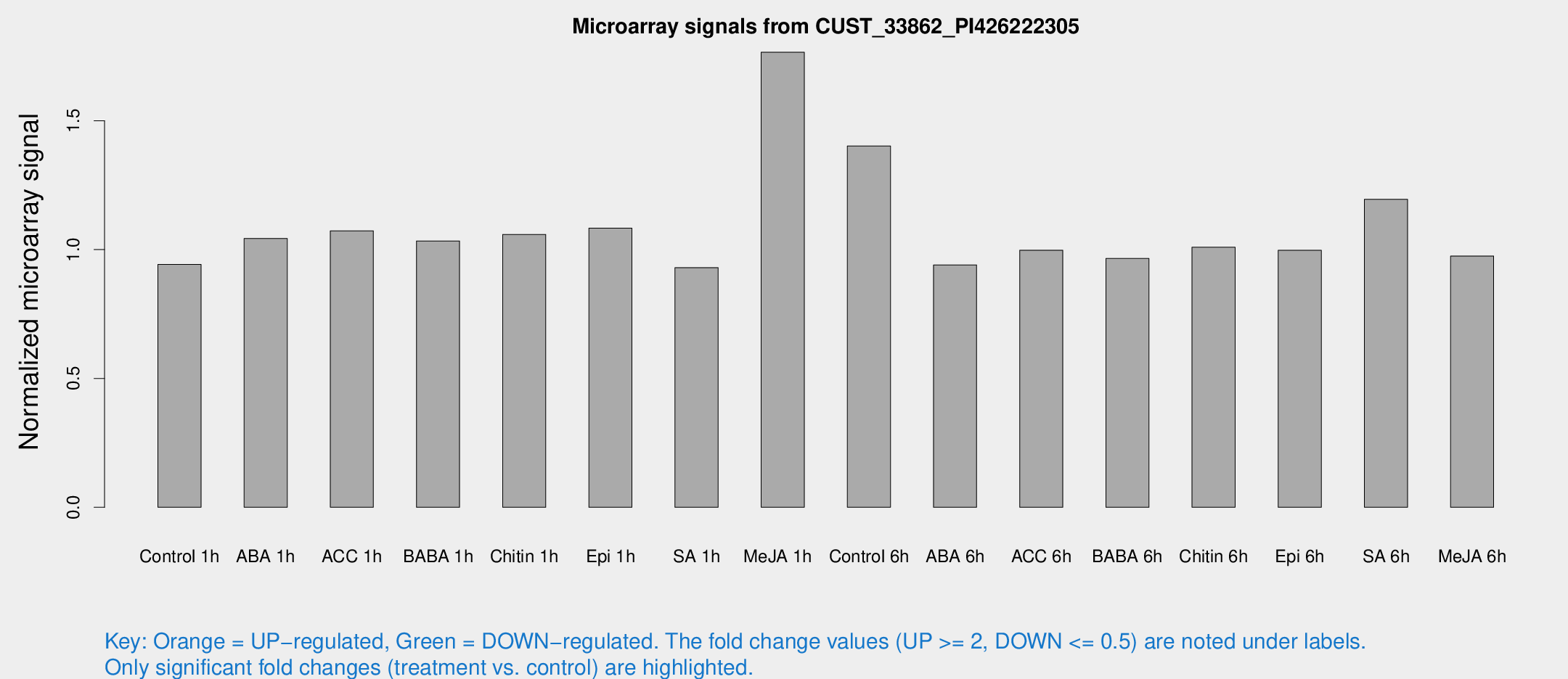
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 5.32586 | 3.08997 | 0.942524 | 0.546235 |
| ABA 1h | 5.15912 | 2.99121 | 1.04332 | 0.597695 |
| ACC 1h | 6.31049 | 3.71116 | 1.07274 | 0.621194 |
| BABA 1h | 5.63118 | 3.26518 | 1.03364 | 0.598536 |
| Chitin 1h | 5.30519 | 3.08363 | 1.05893 | 0.603569 |
| Epi 1h | 5.24305 | 3.04171 | 1.0834 | 0.620552 |
| SA 1h | 5.39594 | 3.12626 | 0.930153 | 0.53865 |
| Me-JA 1h | 8.65426 | 3.23596 | 1.7659 | 0.737201 |
| Control 6h | 8.31167 | 3.18787 | 1.40213 | 0.596266 |
| ABA 6h | 5.64963 | 3.27501 | 0.940452 | 0.544591 |
| ACC 6h | 6.61518 | 3.9271 | 0.997761 | 0.577783 |
| BABA 6h | 6.11304 | 3.54851 | 0.965686 | 0.559547 |
| Chitin 6h | 6.06542 | 3.51296 | 1.00919 | 0.584347 |
| Epi 6h | 6.39101 | 3.72813 | 0.997178 | 0.577564 |
| SA 6h | 6.86949 | 3.37447 | 1.1953 | 0.616048 |
| Me-JA 6h | 5.48537 | 3.17925 | 0.974981 | 0.564572 |
Source Transcript PGSC0003DMT400012376 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G72880.2 | +2 | 4e-102 | 308 | 158/242 (65%) | Survival protein SurE-like phosphatase/nucleotidase | chr1:27423678-27425928 REVERSE LENGTH=385 |
