Probe CUST_33837_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_33837_PI426222305 | JHI_St_60k_v1 | DMT400012378 | TTTTATGTGACAGGGATTCAAATCGACCAGGCAAAGTCATTGGAGCCCTAAGCTTTGCTG |
All Microarray Probes Designed to Gene DMG402004850
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_33862_PI426222305 | JHI_St_60k_v1 | DMT400012376 | TTCCTTGGTCAGACCAGTTCATCAAAAAGAGTCTCCATTCGAAACTGCTAATTGTCAATC |
| CUST_33937_PI426222305 | JHI_St_60k_v1 | DMT400012377 | CAGCCAGATGAAATATAGTGTAATGTTGTTTGTTACCTGATATGCTAAAAAGTTGAGTGC |
| CUST_33837_PI426222305 | JHI_St_60k_v1 | DMT400012378 | TTTTATGTGACAGGGATTCAAATCGACCAGGCAAAGTCATTGGAGCCCTAAGCTTTGCTG |
Microarray Signals from CUST_33837_PI426222305
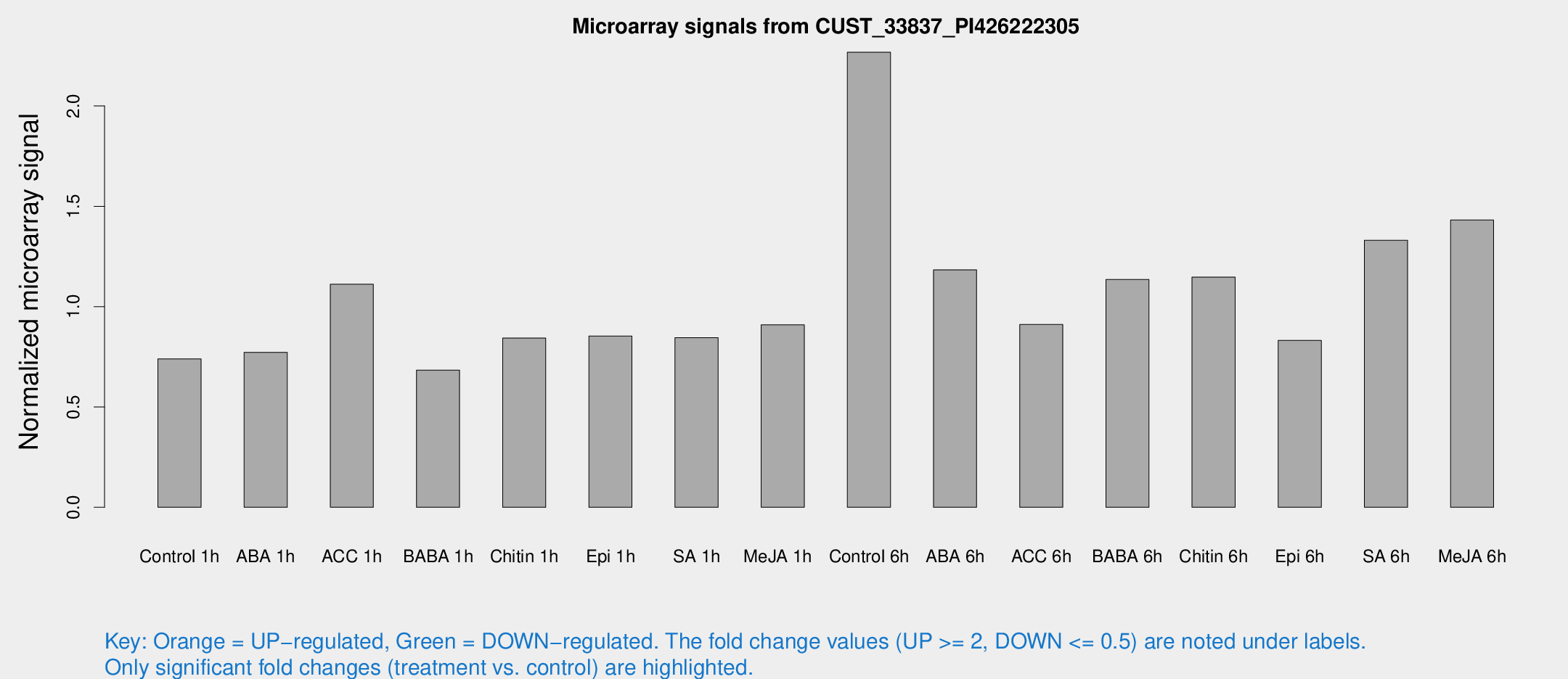
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 8.49888 | 3.46399 | 0.739736 | 0.339485 |
| ABA 1h | 7.64957 | 3.32212 | 0.772291 | 0.350375 |
| ACC 1h | 12.664 | 4.45816 | 1.11158 | 0.419048 |
| BABA 1h | 7.3104 | 3.68448 | 0.683736 | 0.353778 |
| Chitin 1h | 9.61076 | 3.80369 | 0.84346 | 0.378693 |
| Epi 1h | 8.86666 | 3.35213 | 0.853035 | 0.39983 |
| SA 1h | 10.0681 | 3.50889 | 0.844887 | 0.33506 |
| Me-JA 1h | 8.28412 | 3.57495 | 0.909248 | 0.400868 |
| Control 6h | 25.7768 | 4.56017 | 2.2678 | 0.38032 |
| ABA 6h | 13.9653 | 3.74781 | 1.18328 | 0.327806 |
| ACC 6h | 13.6971 | 5.69575 | 0.911039 | 0.464511 |
| BABA 6h | 15.2324 | 4.06864 | 1.13508 | 0.427451 |
| Chitin 6h | 13.4293 | 4.00356 | 1.1471 | 0.345979 |
| Epi 6h | 11.722 | 4.34606 | 0.831867 | 0.387386 |
| SA 6h | 16.4249 | 5.06552 | 1.33054 | 0.686975 |
| Me-JA 6h | 17.3731 | 4.9813 | 1.43151 | 0.705025 |
Source Transcript PGSC0003DMT400012378 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G72880.2 | +3 | 4e-23 | 97 | 88/143 (62%) | Survival protein SurE-like phosphatase/nucleotidase | chr1:27423678-27425928 REVERSE LENGTH=385 |
