Probe CUST_30029_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_30029_PI426222305 | JHI_St_60k_v1 | DMT400005622 | AAATATCTTCTTTGTAATGGTTACTGTGAGAAGTTTTGAATCCATCTGTACATGCTGGAC |
All Microarray Probes Designed to Gene DMG400002204
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_30027_PI426222305 | JHI_St_60k_v1 | DMT400005624 | AAATATCTTCTTTGTAATGGTTACTGTGAGAAGTTTTGAATCCATCTGTACATGCTGGAC |
| CUST_30029_PI426222305 | JHI_St_60k_v1 | DMT400005622 | AAATATCTTCTTTGTAATGGTTACTGTGAGAAGTTTTGAATCCATCTGTACATGCTGGAC |
| CUST_30016_PI426222305 | JHI_St_60k_v1 | DMT400005623 | AAATATCTTCTTTGTAATGGTTACTGTGAGAAGTTTTGAATCCATCTGTACATGCTGGAC |
| CUST_30001_PI426222305 | JHI_St_60k_v1 | DMT400005621 | AAATATCTTCTTTGTAATGGTTACTGTGAGAAGTTTTGAATCCATCTGTACATGCTGGAC |
| CUST_29983_PI426222305 | JHI_St_60k_v1 | DMT400005620 | AAATATCTTCTTTGTAATGGTTACTGTGAGAAGTTTTGAATCCATCTGTACATGCTGGAC |
Microarray Signals from CUST_30029_PI426222305
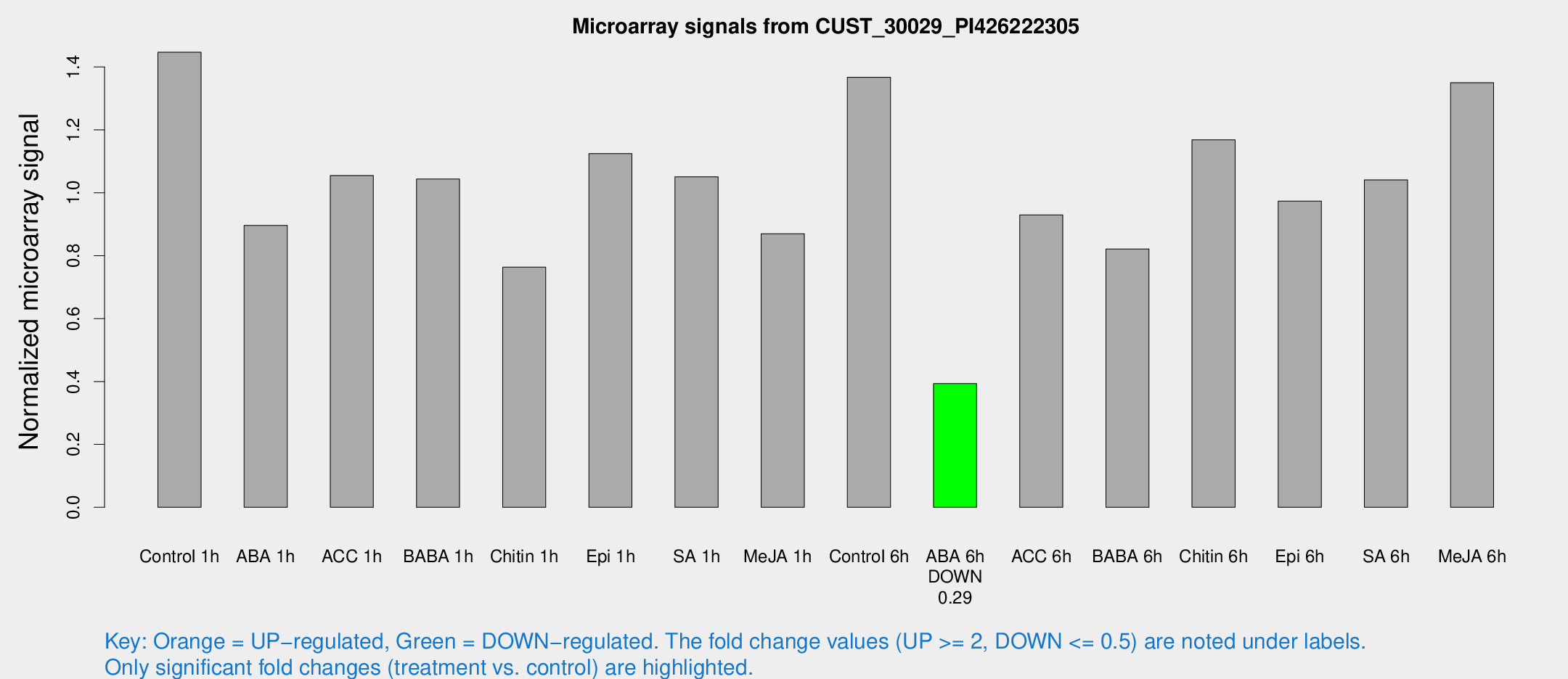
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 1128.99 | 181.918 | 1.4469 | 0.135072 |
| ABA 1h | 602.984 | 34.957 | 0.896111 | 0.0543229 |
| ACC 1h | 934.617 | 281.161 | 1.0549 | 0.342811 |
| BABA 1h | 765.881 | 44.395 | 1.04367 | 0.0835873 |
| Chitin 1h | 531.549 | 70.8044 | 0.763518 | 0.0919016 |
| Epi 1h | 742.489 | 43.1094 | 1.12463 | 0.0793711 |
| SA 1h | 848.893 | 161.463 | 1.05094 | 0.197633 |
| Me-JA 1h | 540.595 | 31.4201 | 0.869801 | 0.0638442 |
| Control 6h | 1123.43 | 272.187 | 1.36716 | 0.278389 |
| ABA 6h | 320.195 | 25.2759 | 0.393395 | 0.033139 |
| ACC 6h | 821.146 | 90.0379 | 0.929899 | 0.0538684 |
| BABA 6h | 720.805 | 126.656 | 0.821558 | 0.12799 |
| Chitin 6h | 948.534 | 55.0286 | 1.16862 | 0.0939778 |
| Epi 6h | 861.789 | 143.547 | 0.973649 | 0.260292 |
| SA 6h | 809.481 | 151.383 | 1.04084 | 0.127055 |
| Me-JA 6h | 1039.62 | 130.399 | 1.3498 | 0.0780643 |
Source Transcript PGSC0003DMT400005622 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G56750.1 | +1 | 4e-135 | 372 | 190/259 (73%) | N-MYC downregulated-like 1 | chr5:22957986-22960606 FORWARD LENGTH=346 |
