Probe CUST_30016_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_30016_PI426222305 | JHI_St_60k_v1 | DMT400005623 | AAATATCTTCTTTGTAATGGTTACTGTGAGAAGTTTTGAATCCATCTGTACATGCTGGAC |
All Microarray Probes Designed to Gene DMG400002204
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_30027_PI426222305 | JHI_St_60k_v1 | DMT400005624 | AAATATCTTCTTTGTAATGGTTACTGTGAGAAGTTTTGAATCCATCTGTACATGCTGGAC |
| CUST_30029_PI426222305 | JHI_St_60k_v1 | DMT400005622 | AAATATCTTCTTTGTAATGGTTACTGTGAGAAGTTTTGAATCCATCTGTACATGCTGGAC |
| CUST_30016_PI426222305 | JHI_St_60k_v1 | DMT400005623 | AAATATCTTCTTTGTAATGGTTACTGTGAGAAGTTTTGAATCCATCTGTACATGCTGGAC |
| CUST_30001_PI426222305 | JHI_St_60k_v1 | DMT400005621 | AAATATCTTCTTTGTAATGGTTACTGTGAGAAGTTTTGAATCCATCTGTACATGCTGGAC |
| CUST_29983_PI426222305 | JHI_St_60k_v1 | DMT400005620 | AAATATCTTCTTTGTAATGGTTACTGTGAGAAGTTTTGAATCCATCTGTACATGCTGGAC |
Microarray Signals from CUST_30016_PI426222305
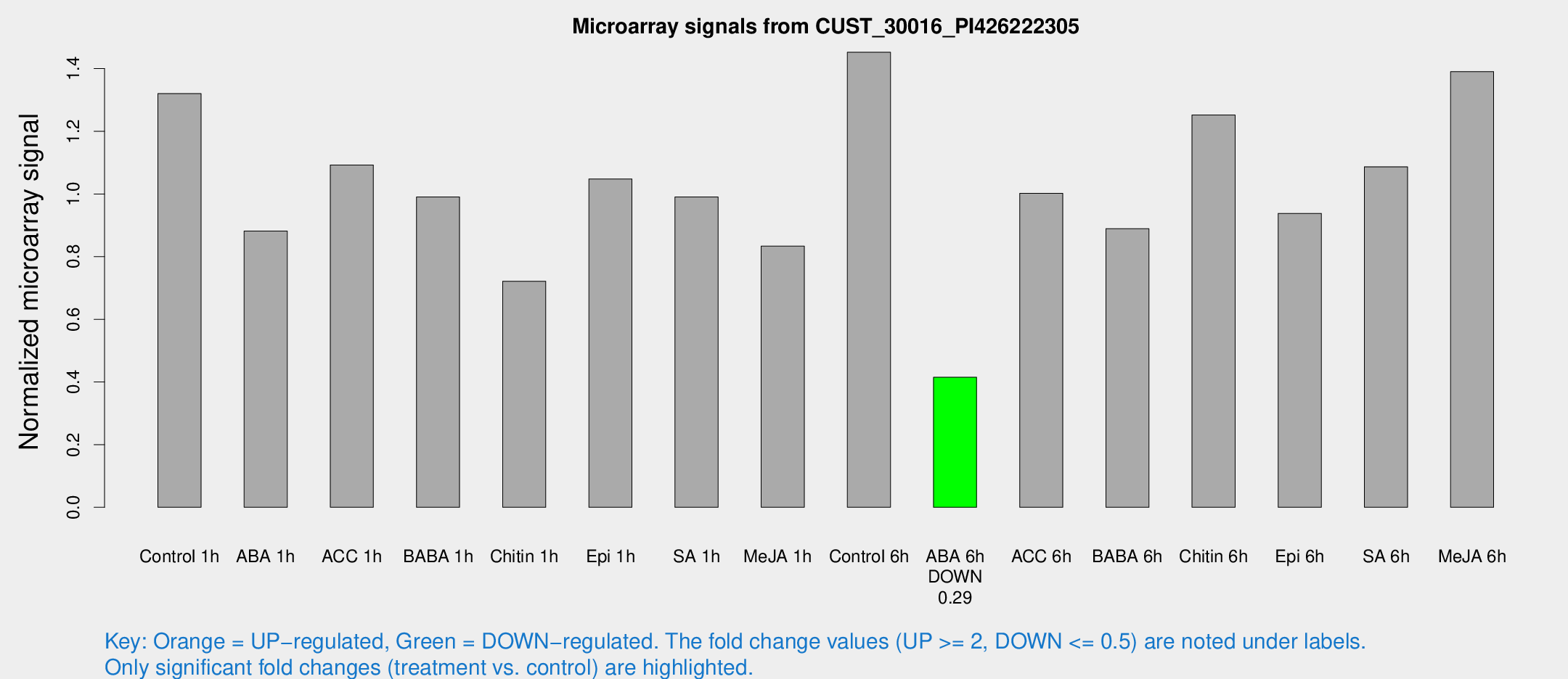
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 1079.54 | 223.775 | 1.32011 | 0.181323 |
| ABA 1h | 613.275 | 35.5436 | 0.881476 | 0.0599162 |
| ACC 1h | 954.284 | 239.806 | 1.09264 | 0.243531 |
| BABA 1h | 754.26 | 51.7492 | 0.990469 | 0.0573444 |
| Chitin 1h | 518.061 | 64.3941 | 0.721541 | 0.0812651 |
| Epi 1h | 718.639 | 60.8594 | 1.04803 | 0.0756758 |
| SA 1h | 835.12 | 179.799 | 0.990302 | 0.230554 |
| Me-JA 1h | 536.84 | 31.2587 | 0.833647 | 0.0483786 |
| Control 6h | 1228.52 | 289.729 | 1.45235 | 0.281219 |
| ABA 6h | 351.091 | 37.437 | 0.415149 | 0.040395 |
| ACC 6h | 908.659 | 66.5288 | 1.00207 | 0.0733334 |
| BABA 6h | 797.649 | 107.071 | 0.889266 | 0.0997476 |
| Chitin 6h | 1048.09 | 60.6316 | 1.25188 | 0.0723965 |
| Epi 6h | 854.634 | 132.715 | 0.93809 | 0.22561 |
| SA 6h | 879.765 | 171.999 | 1.08679 | 0.115601 |
| Me-JA 6h | 1095.88 | 89.9985 | 1.39046 | 0.08038 |
Source Transcript PGSC0003DMT400005623 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G56750.1 | +2 | 2e-173 | 495 | 248/331 (75%) | N-MYC downregulated-like 1 | chr5:22957986-22960606 FORWARD LENGTH=346 |
