Probe CUST_29235_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_29235_PI426222305 | JHI_St_60k_v1 | DMT400053140 | GGTTCATAGATATCCTTATTGTTACACAAATGACTCAGATATACCGCTAACTTATGTCGT |
All Microarray Probes Designed to Gene DMG400020618
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_29235_PI426222305 | JHI_St_60k_v1 | DMT400053140 | GGTTCATAGATATCCTTATTGTTACACAAATGACTCAGATATACCGCTAACTTATGTCGT |
| CUST_29241_PI426222305 | JHI_St_60k_v1 | DMT400053141 | GTTAACGCTGTTGCACCTTGGATTATTGATACCGCCCTTACAGATACTGTAGCTGAAGAT |
Microarray Signals from CUST_29235_PI426222305
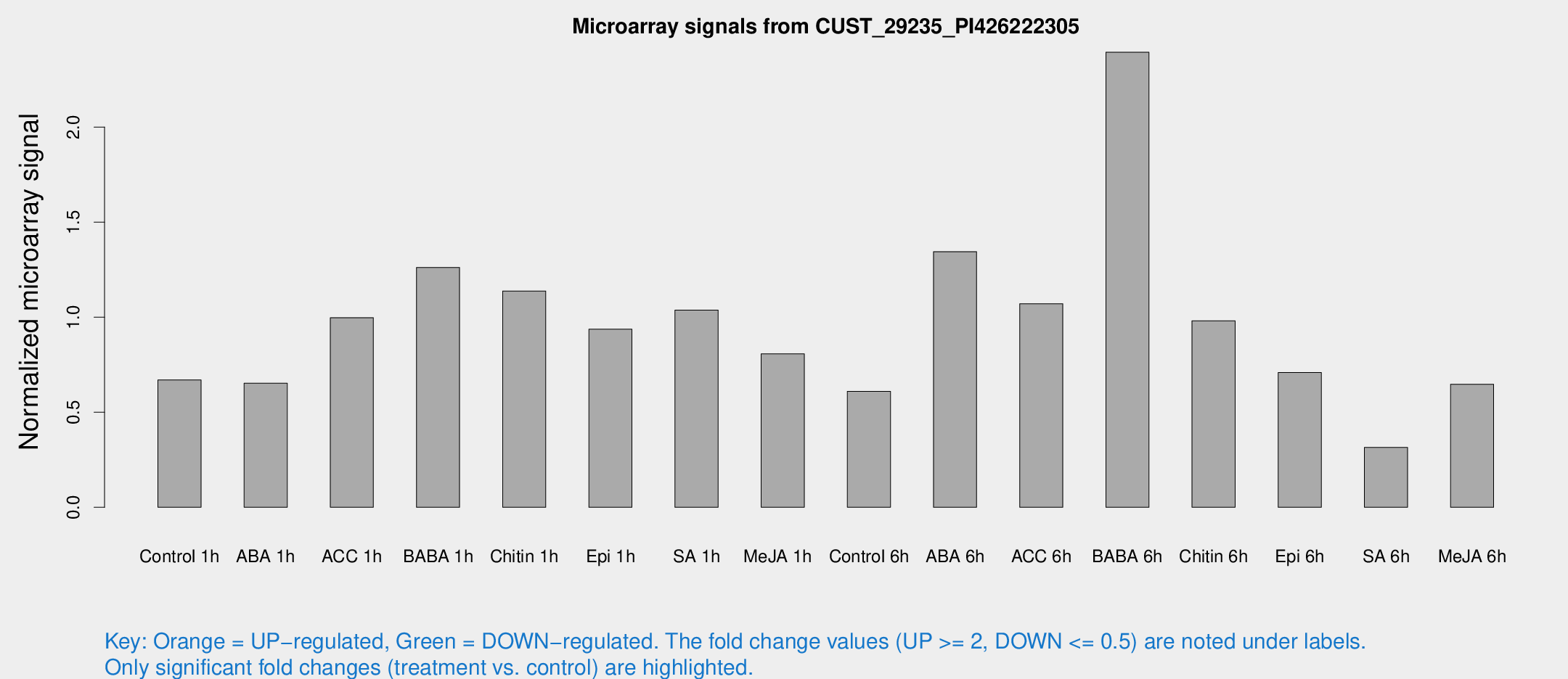
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 26.7043 | 12.1819 | 0.670293 | 0.405551 |
| ABA 1h | 18.9346 | 3.29884 | 0.652894 | 0.117014 |
| ACC 1h | 39.6893 | 14.4884 | 0.997436 | 0.421782 |
| BABA 1h | 41.2472 | 8.31721 | 1.26172 | 0.158391 |
| Chitin 1h | 33.3105 | 3.80407 | 1.1376 | 0.183415 |
| Epi 1h | 27.6853 | 6.04657 | 0.936618 | 0.21181 |
| SA 1h | 40.6014 | 13.3112 | 1.03768 | 0.633705 |
| Me-JA 1h | 23.7675 | 7.6604 | 0.807009 | 0.257687 |
| Control 6h | 28.3736 | 15.302 | 0.61033 | 0.421819 |
| ABA 6h | 49.2715 | 11.8573 | 1.34476 | 0.280196 |
| ACC 6h | 40.3005 | 4.76271 | 1.07101 | 0.12431 |
| BABA 6h | 101.156 | 40.1189 | 2.39371 | 0.925393 |
| Chitin 6h | 34.0235 | 4.36754 | 0.981237 | 0.12821 |
| Epi 6h | 33.3529 | 14.8655 | 0.709189 | 0.541834 |
| SA 6h | 10.608 | 3.72157 | 0.314917 | 0.125034 |
| Me-JA 6h | 26.4486 | 10.6109 | 0.647513 | 0.343009 |
Source Transcript PGSC0003DMT400053140 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G06060.1 | +1 | 3e-72 | 232 | 110/199 (55%) | NAD(P)-binding Rossmann-fold superfamily protein | chr5:1824066-1825833 REVERSE LENGTH=264 |
