Probe CUST_29241_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_29241_PI426222305 | JHI_St_60k_v1 | DMT400053141 | GTTAACGCTGTTGCACCTTGGATTATTGATACCGCCCTTACAGATACTGTAGCTGAAGAT |
All Microarray Probes Designed to Gene DMG400020618
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_29235_PI426222305 | JHI_St_60k_v1 | DMT400053140 | GGTTCATAGATATCCTTATTGTTACACAAATGACTCAGATATACCGCTAACTTATGTCGT |
| CUST_29241_PI426222305 | JHI_St_60k_v1 | DMT400053141 | GTTAACGCTGTTGCACCTTGGATTATTGATACCGCCCTTACAGATACTGTAGCTGAAGAT |
Microarray Signals from CUST_29241_PI426222305
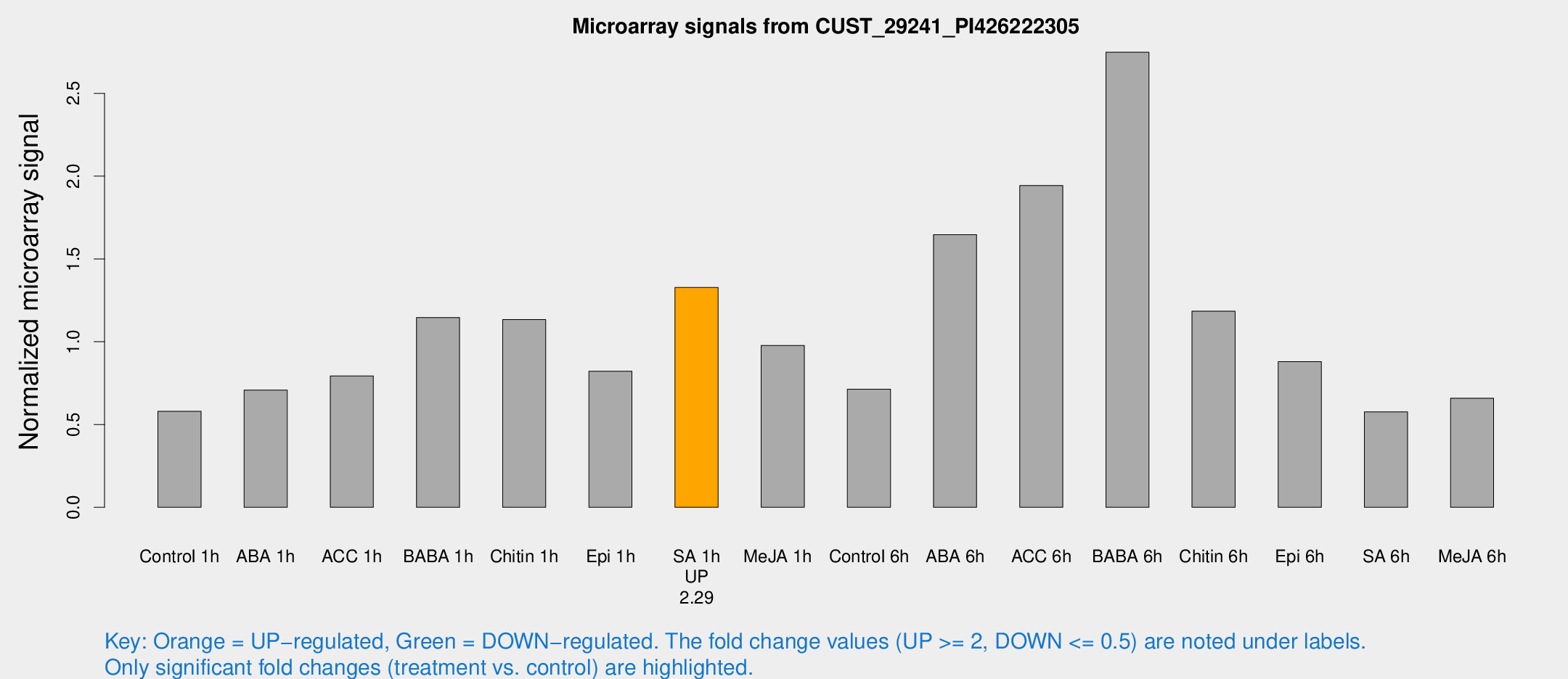
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 2216.46 | 128.119 | 0.579339 | 0.0334593 |
| ABA 1h | 2555.94 | 653.566 | 0.708353 | 0.133928 |
| ACC 1h | 3506.42 | 1082.34 | 0.793589 | 0.239783 |
| BABA 1h | 4512.94 | 1090.07 | 1.1464 | 0.196515 |
| Chitin 1h | 3899.45 | 225.315 | 1.13369 | 0.106711 |
| Epi 1h | 2727.98 | 157.988 | 0.822233 | 0.0474823 |
| SA 1h | 5228.61 | 302.778 | 1.3276 | 0.0766539 |
| Me-JA 1h | 3151.45 | 587.541 | 0.977858 | 0.0979657 |
| Control 6h | 3179.53 | 1035.95 | 0.7138 | 0.251145 |
| ABA 6h | 6800.82 | 846.257 | 1.64654 | 0.123893 |
| ACC 6h | 8617.17 | 874.717 | 1.94358 | 0.231649 |
| BABA 6h | 12114.1 | 2153.29 | 2.74841 | 0.417362 |
| Chitin 6h | 4878.08 | 514.388 | 1.1847 | 0.163305 |
| Epi 6h | 3859.06 | 497.963 | 0.880199 | 0.16235 |
| SA 6h | 2391.77 | 674.77 | 0.576625 | 0.122242 |
| Me-JA 6h | 2995.29 | 1025.7 | 0.658845 | 0.281759 |
Source Transcript PGSC0003DMT400053141 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G06060.1 | +1 | 7e-99 | 298 | 145/257 (56%) | NAD(P)-binding Rossmann-fold superfamily protein | chr5:1824066-1825833 REVERSE LENGTH=264 |
